TATA box-binding protein (TBP)-related factor 2 (TRF2), a third member of the TBP family - PubMed (original) (raw)
TATA box-binding protein (TBP)-related factor 2 (TRF2), a third member of the TBP family
M D Rabenstein et al. Proc Natl Acad Sci U S A. 1999.
Abstract
The TATA box-binding protein (TBP) is an essential component of the RNA polymerase II transcription apparatus in eukaryotic cells. Until recently, it was thought that the general transcriptional machinery was largely invariant and relied on a single TBP, whereas a large and diverse collection of activators and repressors were primarily responsible for imparting specificity to transcription initiation. However, it now appears that the "basal" transcriptional machinery also contributes to specificity via tissue-specific versions of TBP-associated factors as well as a tissue-specific TBP-related factor (TRF1) responsible for gene selectivity in Drosophila. Here we report the cloning of a TBP-related factor (TRF2) that is found in humans, Drosophila, Caenorhabditis elegans, and other metazoans. Like TRF1 and TBP, TRF2 binds transcription factor IIA (TFIIA) and TFIIB and appears to be part of a larger protein complex. TRF2's primary amino acid structure suggests divergence in the putative DNA binding domain, and not surprisingly, it fails to bind to DNA containing canonical TATA boxes. Most importantly, TRF2 is associated with loci on Drosophila chromosomes distinct from either TBP or TRF1, so it may have different promoter specificity and regulate a select subset of genes. These findings suggest that metazoans have evolved multiple TBPs to accommodate the vast increase in genes and expression patterns during development and cellular differentiation.
Figures
Figure 1
Comparison of TBP family members. (a) Percent identity in the bipartite repeat domain between the Drosophila TBP family members. (b) Organization of domains in TBP family members. The arrows represent the bipartite repeats of the core domain. (c) Alignment of the core domains of dTRF2, hTRF2, ceTRF2, and the TBP consensus sequence, with “X” denoting nonconserved residues. ∗ denotes residues conserved between TRF2 family members that are different from the TBP consensus. The TBP consensus was obtained by comparing Drosophila, human, and C. elegans TBP. Shown are residues 198–379 of dTRF2, 7–186 of hTRF2, and 178–466 of C. elegans TRF2. Full-length Drosophila TRF2 and full-lentgh human TRF2 have been deposited in the GenBank database (accession nos. AF136569 and AF136570). # in the ceTRF2 sequence denotes the 109-residue insertion.
Figure 2
Analysis of dTBP, dTRF1, and dTRF2 conserved residues. (a) Alignment of dTBP, dTRF1, and dTRF2. Red boxes highlight identical residues; a, b, and d indicate TFIIA-, TFIIB-, and DNA-binding residues, respectively (18, 23, 25, 26). (b and c) Location of residues conserved in dTRF1 (a) and dTRF2 (b) visualized on the TBP model. Red residues are conserved with TBP, and blue residues are nonconserved. The TBP model is as described in ref. , with the N-terminal repeat on the right.
Figure 3
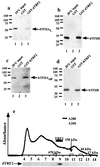
Biochemical analysis of TRF2. (a_–_d) Western blot analysis of TFIIA and TFIIB binding to TRF2. A GST fusion of dTRF2 (a and b) or hTRF2 (c and d) and GST (a_–_d) was bound to glutathione beads and then incubated with dTFIIA (a), dTFIIB (b), hTFIIA (c), or hTFIIB (d). Unbound protein was then washed away, and the bound protein was detected by Western blot analysis. (e) Elution of endogenous dTRF2 from a Superose 6 gel filtration column (Amersham Pharmacia). Endogenous Drosophila TRF2 eluted slightly after the 670-kDa molecular mass standard. The elution profile of the molecular mass standards is indicated along the axis.
Figure 4
Northern blot analysis of hTRF2 expression in multiple human tissues. (Upper) Blots were probed with TRF2 and actin. An alternative TRF2 transcript was observed in testis (∗), and an alternative actin transcript was observed in both muscle and heart (∗). (Lower) Comparison of relative hTRF2 levels. The hTRF2 signal was normalized against actin, and the testis level was arbitrarily set to 100. pbl, peripheral blood leukocyte.
Figure 5
TRF2 and TAFII250 localization on Drosophila polytene chromosomes. Anti-TAFII250 (raised in a mouse) antibodies visualized with Texas red-conjugated secondary antibody is shown in red, and anti-dTRF2 (raised in a rabbit) antibodies visualized with FITC-conjugated secondary antibodies is shown in green. (a) Anti-TAFII250 only. (b) Both anti-TAFII250 and anti-dTRF2. (c) Anti-dTRF2 only.
Similar articles
- Role of the TATA-box binding protein (TBP) and associated family members in transcription regulation.
Mishal R, Luna-Arias JP. Mishal R, et al. Gene. 2022 Jul 30;833:146581. doi: 10.1016/j.gene.2022.146581. Epub 2022 May 18. Gene. 2022. PMID: 35597524 Review. - A new factor related to TATA-binding protein has highly restricted expression patterns in Drosophila.
Crowley TE, Hoey T, Liu JK, Jan YN, Jan LY, Tjian R. Crowley TE, et al. Nature. 1993 Feb 11;361(6412):557-61. doi: 10.1038/361557a0. Nature. 1993. PMID: 8429912 - Human TATA-binding protein-related factor-2 (hTRF2) stably associates with hTFIIA in HeLa cells.
Teichmann M, Wang Z, Martinez E, Tjernberg A, Zhang D, Vollmer F, Chait BT, Roeder RG. Teichmann M, et al. Proc Natl Acad Sci U S A. 1999 Nov 23;96(24):13720-5. doi: 10.1073/pnas.96.24.13720. Proc Natl Acad Sci U S A. 1999. PMID: 10570139 Free PMC article. - TRF2: TRansForming the view of general transcription factors.
Zehavi Y, Kedmi A, Ideses D, Juven-Gershon T. Zehavi Y, et al. Transcription. 2015;6(1):1-6. doi: 10.1080/21541264.2015.1004980. Epub 2015 Jan 14. Transcription. 2015. PMID: 25588059 Free PMC article. Review. - Identification of a mouse TBP-like protein (TLP) distantly related to the drosophila TBP-related factor.
Ohbayashi T, Makino Y, Tamura TA. Ohbayashi T, et al. Nucleic Acids Res. 1999 Feb 1;27(3):750-5. doi: 10.1093/nar/27.3.750. Nucleic Acids Res. 1999. PMID: 9889269 Free PMC article.
Cited by
- Specialized and redundant roles of TBP and a vertebrate-specific TBP paralog in embryonic gene regulation in Xenopus.
Jallow Z, Jacobi UG, Weeks DL, Dawid IB, Veenstra GJ. Jallow Z, et al. Proc Natl Acad Sci U S A. 2004 Sep 14;101(37):13525-30. doi: 10.1073/pnas.0405536101. Epub 2004 Sep 2. Proc Natl Acad Sci U S A. 2004. PMID: 15345743 Free PMC article. - Drosophila TRF2 is a preferential core promoter regulator.
Kedmi A, Zehavi Y, Glick Y, Orenstein Y, Ideses D, Wachtel C, Doniger T, Waldman Ben-Asher H, Muster N, Thompson J, Anderson S, Avrahami D, Yates JR 3rd, Shamir R, Gerber D, Juven-Gershon T. Kedmi A, et al. Genes Dev. 2014 Oct 1;28(19):2163-74. doi: 10.1101/gad.245670.114. Epub 2014 Sep 15. Genes Dev. 2014. PMID: 25223897 Free PMC article. - A transcription factor IIA-binding site differentially regulates RNA polymerase II-mediated transcription in a promoter context-dependent manner.
Wang J, Zhao S, He W, Wei Y, Zhang Y, Pegg H, Shore P, Roberts SGE, Deng W. Wang J, et al. J Biol Chem. 2017 Jul 14;292(28):11873-11885. doi: 10.1074/jbc.M116.770412. Epub 2017 May 24. J Biol Chem. 2017. PMID: 28539359 Free PMC article. - A sequence-specific core promoter-binding transcription factor recruits TRF2 to coordinately transcribe ribosomal protein genes.
Baumann DG, Gilmour DS. Baumann DG, et al. Nucleic Acids Res. 2017 Oct 13;45(18):10481-10491. doi: 10.1093/nar/gkx676. Nucleic Acids Res. 2017. PMID: 28977400 Free PMC article. - Characterizing dopaminergic neuron vulnerability using genome-wide analysis.
Davis J, Da Silva Santos C, Zavala NC, Gans N, Patracuolla D, Fehrenbach M, Babcock DT. Davis J, et al. Genetics. 2021 Aug 9;218(4):iyab081. doi: 10.1093/genetics/iyab081. Genetics. 2021. PMID: 34038543 Free PMC article.
References
- Tjian R, Maniatis T. Cell. 1994;77:5–8. - PubMed
- Goodrich J A, Cutler G, Tjian R. Cell. 1996;84:825–830. - PubMed
- Dikstein R, Zhou S, Tjian R. Cell. 1996;87:137–146. - PubMed
- Jacq X, Brou C, Lutz Y, Davidson I, Chambon P, Tora L. Cell. 1994;79:107–117. - PubMed
- Grant P A, Schieltz D, Pray-Grant M G, Steger D J, Reese J C, Yates J R, III, Workman J L. Cell. 1998;94:45–53. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- GM25232/GM/NIGMS NIH HHS/United States
- R37 CA025417/CA/NCI NIH HHS/United States
- R37 GM025232/GM/NIGMS NIH HHS/United States
- CA25417/CA/NCI NIH HHS/United States
- R01 GM025232/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous