Three proteins define a class of human histone deacetylases related to yeast Hda1p - PubMed (original) (raw)
Three proteins define a class of human histone deacetylases related to yeast Hda1p
C M Grozinger et al. Proc Natl Acad Sci U S A. 1999.
Abstract
Gene expression is in part controlled by chromatin remodeling factors and the acetylation state of nucleosomal histones. The latter process is regulated by histone acetyltransferases and histone deacetylases (HDACs). Previously, three human and five yeast HDAC enzymes had been identified. These can be categorized into two classes: the first class represented by yeast Rpd3-like proteins and the second by yeast Hda1-like proteins. Human HDAC1, HDAC2, and HDAC3 proteins are members of the first class, whereas no class II human HDAC proteins had been identified. The amino acid sequence of Hda1p was used to search the GenBank/expressed sequence tag databases to identify partial sequences from three putative class II human HDAC proteins. The corresponding full-length cDNAs were cloned and defined as HDAC4, HDAC5, and HDAC6. These proteins possess certain features present in the conserved catalytic domains of class I human HDACs, but also contain additional sequence domains. Interestingly, HDAC6 contains an internal duplication of two catalytic domains, which appear to function independently of each other. These class II HDAC proteins have differential mRNA expression in human tissues and possess in vitro HDAC activity that is inhibited by trichostatin A. Coimmunoprecipitation experiments indicate that these HDAC proteins are not components of the previously identified HDAC1 and HDAC2 NRD and mSin3A complexes. However, HDAC4 and HDAC5 associate with HDAC3 in vivo. This finding suggests that the human class II HDAC enzymes may function in cellular processes distinct from those of HDAC1 and HDAC2.
Figures
Figure 1
Predicted amino acid sequences of human class II HDACs. The conserved residues of the catalytic domains are highlighted. (A) HDAC4, (B) HDAC5, and (C) HDAC6 predicted amino acid sequences. Note that there are two putative catalytic domains in HDAC6. (D) Alignment of catalytic domains of yeast HDA1p, human HDAC1, HDAC4, and HDAC5 with both catalytic regions of HDAC6. The residues that are conserved in these HDACs as well as in acuC (B. subtilis, GenBank accession no. 348052) and ASD (M. ramosa, GenBank accession no. 3023317) are in bold type, and those residues that are conserved within the class II human HDAC enzymes are boxed.
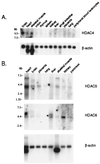
Figure 2
Expression analysis of human class II HDAC family members. Multiple human tissue Northern blots were probed to determine mRNA expression of HDAC4, HDAC5, and HDAC6. Blots were stripped and reprobed with β-actin cDNA to normalize for total mRNA. The position of molecular size markers is indicated on the left.
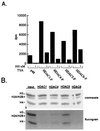
Figure 3
Class II HDAC enzymes deacetylate all four core histones in vitro. Recombinant FLAG-tagged HDACs were immunoprecipitated from transfected Jurkat cell extracts by using α-FLAG antibody (Sigma). Immunopurified enzymes were incubated with radiolabeled core histones as described in Materials and Methods. (A) The HDAC activity was measured by scintillation counting of the released [3H]acetic acid. Where indicated, immunoprecipitates were preincubated with trichostatin A (Wako) before addition of histones. Each assay was performed in duplicate and averaged. (B) Substrate specificity of class II HDACs. Deacetylase reactions were separated by 20% SDS/PAGE and stained with Coomassie (Upper). The gel was treated with EnHance (National Diagnostics), dried, and exposed to film (Lower). The identities of the core histones are indicated on the left. RbAp48 was transfected as a negative control.
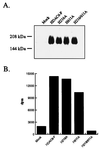
Figure 4
The catalytic domains of HDAC6 function independently. The histidine residues homologous to H141 of HDAC1 in each of the catalytic domains (H216 and H611) were mutated to alanine by PCR overlap extension. The single and double mutants were FLAG-tagged and expressed in Tag-Jurkat cells. The enzymes were immunoprecipitated by using α-FLAG antibodies (Sigma), and expression levels were compared by Western blotting (A). The mutant enzymes then were assayed for HDAC activity as before (B).
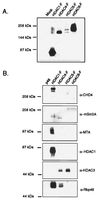
Figure 5
Class II HDAC enzymes and HDAC1 are in different complexes in vivo. Recombinant FLAG-tagged HDACs were precipitated from transfected Jurkat cell extracts by using α-FLAG antibody (Sigma), separated by SDS/PAGE, and subjected to Western blot analysis. Blots were probed with (A) α-FLAG antibody (Sigma) to determine expression levels and (B) α-CHD4, -mSin3A, -MTA, -HDAC1, -HDAC3, and -Rbp48 antibodies to determine whether these proteins coimmunoprecipitated with the class II HDAC enzymes.
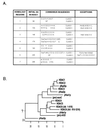
Figure 6
Sequence analysis suggests that HDAC enzymes have diverged into two classes. (A) Alignment of human HDAC enzymes 1–6 with yeast Rpd3p, Hos1p, Hos2p, Hos3p, M. ramosa ASD, and B. subtilis acuC reveals the presence of seven conserved regions, whose consensus sequences differ between the two classes. Amino acids are represented by single letter codes; X represents any amino acid while Φ indicates a hydrophobic residue. NF, not found. (B) A phylogenetic analysis suggests that the HDAC enzymes diverged from a common prokaryotic ancestor to form two classes of HDAC proteins. Proteins from three different phyla were examined. Prokaryotic proteins are preceded by (pro), yeast proteins are preceded by y, and human proteins are capitalized. Note that yHos3p does not correlate well with either HDAC class.
Similar articles
- Class II histone deacetylases: structure, function, and regulation.
Bertos NR, Wang AH, Yang XJ. Bertos NR, et al. Biochem Cell Biol. 2001;79(3):243-52. Biochem Cell Biol. 2001. PMID: 11467738 Review. - A new family of human histone deacetylases related to Saccharomyces cerevisiae HDA1p.
Fischle W, Emiliani S, Hendzel MJ, Nagase T, Nomura N, Voelter W, Verdin E. Fischle W, et al. J Biol Chem. 1999 Apr 23;274(17):11713-20. doi: 10.1074/jbc.274.17.11713. J Biol Chem. 1999. PMID: 10206986 - The histone deacetylase-3 complex contains nuclear receptor corepressors.
Wen YD, Perissi V, Staszewski LM, Yang WM, Krones A, Glass CK, Rosenfeld MG, Seto E. Wen YD, et al. Proc Natl Acad Sci U S A. 2000 Jun 20;97(13):7202-7. doi: 10.1073/pnas.97.13.7202. Proc Natl Acad Sci U S A. 2000. PMID: 10860984 Free PMC article. - Analysis of histone acetyltransferase and histone deacetylase families of Arabidopsis thaliana suggests functional diversification of chromatin modification among multicellular eukaryotes.
Pandey R, Müller A, Napoli CA, Selinger DA, Pikaard CS, Richards EJ, Bender J, Mount DW, Jorgensen RA. Pandey R, et al. Nucleic Acids Res. 2002 Dec 1;30(23):5036-55. doi: 10.1093/nar/gkf660. Nucleic Acids Res. 2002. PMID: 12466527 Free PMC article. - Epigenetic therapy of cancer with histone deacetylase inhibitors.
Lakshmaiah KC, Jacob LA, Aparna S, Lokanatha D, Saldanha SC. Lakshmaiah KC, et al. J Cancer Res Ther. 2014 Jul-Sep;10(3):469-78. doi: 10.4103/0973-1482.137937. J Cancer Res Ther. 2014. PMID: 25313724 Review.
Cited by
- Identification of hsa_circ_0018905 as a New Potential Biomarker for Multiple Sclerosis.
Lodde V, Zarbo IR, Farina G, Masia A, Solla P, Campesi I, Delogu G, Muroni MR, Tsitsipatis D, Gorospe M, Floris M, Idda ML. Lodde V, et al. Cells. 2024 Oct 9;13(19):1668. doi: 10.3390/cells13191668. Cells. 2024. PMID: 39404430 Free PMC article. - PROTACs Targeting Epigenetic Proteins.
Zhang C, He Y, Sun X, Wei W, Liu Y, Rao Y. Zhang C, et al. Acta Mater Med. 2023 Oct 26;2(4):409-429. doi: 10.15212/amm-2023-0039. Epub 2023 Dec 6. Acta Mater Med. 2023. PMID: 39221114 Free PMC article. - Histone deacetylases: potential therapeutic targets for idiopathic pulmonary fibrosis.
Cheng HP, Jiang SH, Cai J, Luo ZQ, Li XH, Feng DD. Cheng HP, et al. Front Cell Dev Biol. 2024 Aug 13;12:1426508. doi: 10.3389/fcell.2024.1426508. eCollection 2024. Front Cell Dev Biol. 2024. PMID: 39193364 Free PMC article. Review. - Histone deacetylase 6 as a novel promising target to treat cardiovascular disease.
Wu YX, Li BQ, Yu XQ, Liu YL, Chui RH, Sun K, Geng DG, Ma LY. Wu YX, et al. Cancer Innov. 2024 May 7;3(3):e114. doi: 10.1002/cai2.114. eCollection 2024 Jun. Cancer Innov. 2024. PMID: 38947757 Free PMC article. Review. - A novel insight into neurological disorders through HDAC6 protein-protein interactions.
Bahram Sangani N, Koetsier J, Mélius J, Kutmon M, Ehrhart F, Evelo CT, Curfs LMG, Reutelingsperger CP, Eijssen LMT. Bahram Sangani N, et al. Sci Rep. 2024 Jun 25;14(1):14666. doi: 10.1038/s41598-024-65094-1. Sci Rep. 2024. PMID: 38918466 Free PMC article.
References
- Luger K, Mader A W, Richmond R K, Sargent D F, Richmond T J. Nature (London) 1997;389:251–260. - PubMed
- Workman J L, Kingston R E. Annu Rev Biochem. 1998;67:545–579. - PubMed
- Varga-Weisz P D, Becker P B. Curr Opin Cell Biol. 1998;10:346–353. - PubMed
- Tong J K, Hassig C A, Schnitzler G R, Kingston R E, Schreiber S L. Nature (London) 1998;395:917–921. - PubMed
- Zhang Y, LeRoy G, Seelig H P, Lane W S, Reinberg D. Cell. 1998;95:279–289. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous