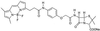BOCILLIN FL, a sensitive and commercially available reagent for detection of penicillin-binding proteins - PubMed (original) (raw)
BOCILLIN FL, a sensitive and commercially available reagent for detection of penicillin-binding proteins
G Zhao et al. Antimicrob Agents Chemother. 1999 May.
Abstract
We describe a new, sensitive, rapid, and nonradioactive method involving the use of the commercially available BOCILLIN FL, a fluorescent penicillin, as a labeling reagent for the detection and study of penicillin-binding proteins (PBPs). This method allowed rapid detection of 30 ng of a purified PBP protein under UV light and of 2 to 4 ng of the protein with the aid of a FluorImager. This method also allowed rapid determination of the PBP profiles of Escherichia coli, Pseudomonas aeruginosa, and Streptococcus pneumoniae. The PBP profiles obtained are virtually identical to those reported previously with 3H-, 14C-, or 125I-labeled penicillin. Using this method enabled us to determine the 50% inhibitory concentrations of the penicillin-sensitive and -resistant PBP2x proteins of S. pneumoniae for penicillin G, thereby allowing a direct evaluation of their relative affinities for penicillin G. Finally, this method also allowed us to compare relative affinities of a PBP2x protein for different beta-lactam antibiotics with the aid of fluorescence polarization technology and to monitor a PBP2x protein during purification.
Figures
FIG. 1
Structure of BOCILLIN FL.
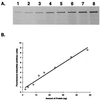
FIG. 2
Detection and quantitation of the penicillin-sensitive PBP2x protein of S. pneumoniae by BOCILLIN FL-binding assays. The penicillin-sensitive PBP2x protein of S. pneumoniae was purified (33), labeled with BOCILLIN FL (10 μM) for 30 min, separated by SDS-PAGE, and visualized by using a FluorImager. (A) Detection of the PBP2x protein; lanes 1 to 8: 2, 4, 8, 12, 16, 24, 36, and 48 ng of the purified PBP2x protein, respectively. (B) Quantitation of the PBP2x protein; fluorescence intensities of each band from panel A were quantified and plotted.
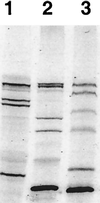
FIG. 3
Detection of PBPs of S. pneumoniae, E. coli, and P. aeruginosa. The membrane fractions of S. pneumoniae, E. coli, and P. aeruginosa were prepared and labeled with BOCILLIN FL (25 μM). The labeled membrane preparations (≈7.5 μg of protein each) were separated by SDS-PAGE and visualized by using a FluorImager. Lanes 1 to 3: BOCILLIN FL-labeled membrane preparations of S. pneumoniae, E. coli, and P. aeruginosa, respectively.
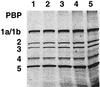
FIG. 4
Minimal amount of BOCILLIN FL required for the detection of E. coli PBPs. The E. coli membrane preparation (≈15 μg of protein each) was labeled with BOCILLIN FL (final concentration, 1.6 to 25 μM). The labeled PBPs are separated by SDS-PAGE and detected by using a FluorImager. Lanes 1 to 5: membrane preparation labeled with 1.6, 3.2, 6.4, 12.5, and 25 μM BOCILLIN FL, respectively.
FIG. 5
Determination of IC50s of the penicillin-sensitive and -resistant PBP2x proteins of S. pneumoniae (hex) R6 and 328, respectively, for penicillin G (Pen G). Both PBP2x proteins were purified (33), labeled with BOCILLIN FL in the presence of various amounts of penicillin G, separated by SDS-PAGE, and visualized by using a FluorImager. (A) Penicillin-resistant PBP2x protein of S. pneumoniae 328. (B) Penicillin-sensitive PBP2x protein of S. pneumoniae (hex) R6.
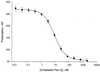
FIG. 6
Determination of the IC50 of the penicillin-sensitive PBP2x protein of S. pneumoniae (hex) R6 for penicillin G (Pen G). The fluorescence polarization for the penicillin-sensitive PBP2x protein (1.3 nM) in a competitive interaction with BOCILLIN FL (2 nM) and increasing concentrations of unlabeled penicillin G (0.01 to 10,000 nM) were measured as described previously. Data points represent the average of four replicates (± standard deviations), and the curve is the predicted nonlinear regression result.
Similar articles
- Fluorescent Bocillins: synthesis and application in the detection of penicillin-binding proteins.
Gee KR, Kang HC, Meier TI, Zhao G, Blaszcak LC. Gee KR, et al. Electrophoresis. 2001 Mar;22(5):960-5. doi: 10.1002/1522-2683()22:5<960::AID-ELPS960>3.0.CO;2-9. Electrophoresis. 2001. PMID: 11332764 - Nucleotide sequences of the pbpX genes encoding the penicillin-binding proteins 2x from Streptococcus pneumoniae R6 and a cefotaxime-resistant mutant, C506.
Laible G, Hakenbeck R, Sicard MA, Joris B, Ghuysen JM. Laible G, et al. Mol Microbiol. 1989 Oct;3(10):1337-48. doi: 10.1111/j.1365-2958.1989.tb00115.x. Mol Microbiol. 1989. PMID: 2615650 - Biological characterization of a new radioactive labeling reagent for bacterial penicillin-binding proteins.
Preston DA, Wu CY, Blaszczak LC, Seitz DE, Halligan NG. Preston DA, et al. Antimicrob Agents Chemother. 1990 May;34(5):718-21. doi: 10.1128/AAC.34.5.718. Antimicrob Agents Chemother. 1990. PMID: 2113792 Free PMC article. - Use of biotinylated beta-lactams and chemiluminescence for study and purification of penicillin-binding proteins in bacteria.
Dargis M, Malouin F. Dargis M, et al. Antimicrob Agents Chemother. 1994 May;38(5):973-80. doi: 10.1128/AAC.38.5.973. Antimicrob Agents Chemother. 1994. PMID: 8067779 Free PMC article. - Penicillin-binding proteins and the mechanism of action of beta-lactam antibiotics.
Waxman DJ, Strominger JL. Waxman DJ, et al. Annu Rev Biochem. 1983;52:825-69. doi: 10.1146/annurev.bi.52.070183.004141. Annu Rev Biochem. 1983. PMID: 6351730 Review. No abstract available.
Cited by
- Characterization of osmotically induced filaments of Salmonella enterica.
Pratt ZL, Chen B, Czuprynski CJ, Wong AC, Kaspar CW. Pratt ZL, et al. Appl Environ Microbiol. 2012 Sep;78(18):6704-13. doi: 10.1128/AEM.01784-12. Epub 2012 Jul 13. Appl Environ Microbiol. 2012. PMID: 22798362 Free PMC article. - First Penicillin-Binding Protein Occupancy Patterns for 15 β-Lactams and β-Lactamase Inhibitors in Mycobacterium abscessus.
Sayed ARM, Shah NR, Basso KB, Kamat M, Jiao Y, Moya B, Sutaria DS, Lang Y, Tao X, Liu W, Shin E, Zhou J, Werkman C, Louie A, Drusano GL, Bulitta JB. Sayed ARM, et al. Antimicrob Agents Chemother. 2020 Dec 16;65(1):e01956-20. doi: 10.1128/AAC.01956-20. Print 2020 Dec 16. Antimicrob Agents Chemother. 2020. PMID: 33106266 Free PMC article. - Expanded profiling of _β_-lactam selectivity for penicillin-binding proteins in Streptococcus pneumoniae D39.
Sharan D, Carlson EE. Sharan D, et al. Biol Chem. 2022 Feb 28;403(4):433-443. doi: 10.1515/hsz-2021-0386. Print 2022 Mar 28. Biol Chem. 2022. PMID: 35218689 Free PMC article. - Genetic and molecular characterization of beta-lactamase-negative ampicillin-resistant Haemophilus influenzae with unusually high resistance to ampicillin.
Kaczmarek FS, Gootz TD, Dib-Hajj F, Shang W, Hallowell S, Cronan M. Kaczmarek FS, et al. Antimicrob Agents Chemother. 2004 May;48(5):1630-9. doi: 10.1128/AAC.48.5.1630-1639.2004. Antimicrob Agents Chemother. 2004. PMID: 15105114 Free PMC article. - Mechanisms of resistance to imipenem and ampicillin in Enterococcus faecalis.
Ono S, Muratani T, Matsumoto T. Ono S, et al. Antimicrob Agents Chemother. 2005 Jul;49(7):2954-8. doi: 10.1128/AAC.49.7.2954-2958.2005. Antimicrob Agents Chemother. 2005. PMID: 15980374 Free PMC article.
References
- Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. - PubMed
- Bush K. Screening and characterization of enzyme inhibitors as drug candidates. Drug Metab Rev. 1983;14:689–708. - PubMed
- Copeland R A. Enzymes: a practical introduction to structure, mechanisms, and data analysis. New York, N.Y: Wiley-VCH; 1996.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous