Export of virulence genes and Shiga toxin by membrane vesicles of Escherichia coli O157:H7 - PubMed (original) (raw)
Export of virulence genes and Shiga toxin by membrane vesicles of Escherichia coli O157:H7
G L Kolling et al. Appl Environ Microbiol. 1999 May.
Abstract
Membrane vesicles released by Escherichia coli O157:H7 into culture medium were purified and analyzed for protein and DNA content. Electron micrographs revealed vesicles that are spherical, range in size from 20 to 100 nm, and have a complete bilayer. Analysis of vesicle protein by sodium dodecyl sulfate-polyacrylamide gel electrophoresis demonstrates vesicles that contain many proteins with molecular sizes similar to outer membrane proteins and a number of cellular proteins. Immunoblot (Western) analysis of vesicles suggests the presence of cell antigens. Treatment of vesicles with exogenous DNase hydrolyzed surface-associated DNA; PCR demonstrated that vesicles contain DNA encoding the virulence genes eae, stx1 and stx2, and uidA, which encodes for beta-galactosidase. Immunoblot analysis of intact and lysed, proteinase K-treated vesicles demonstrate that Shiga toxins 1 and 2 are contained within vesicles. These results suggest that vesicles contain toxic material and transfer experiments demonstrate that vesicles can deliver genetic material to other gram-negative organisms.
Figures
FIG. 1
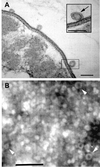
Cells and vesicles of E. coli O157:H7. (A) Ultrathin sections show vesicles associated with a whole cell. The inset is an enlargement of the enclosed area and clearly shows a vesicle membrane bilayer (arrow). Bar = 50 nm. (B) Negatively stained vesicle preparations demonstrate the uniform size and morphology of vesicles. The arrowheads indicate representative individual vesicles. Note that the vesicles appear to contain electron-dense material. Bar = 250 nm.
FIG. 2
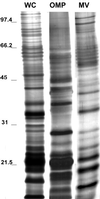
SDS-PAGE protein profiles of whole cells (WC), OMPs, and vesicles (MV) in a 10% polyacrylamide gel stained with silver stain (samples are from strain E. coli O157:H7 ATCC 43895). Each lane contains 25 μg of the total protein. Samples were not heat treated prior to loading. Molecular masses (in kilodaltons) are indicated on the left.
FIG. 3
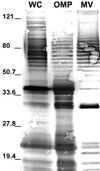
Immunoblot of E. coli O157:H7 whole cells (WC), OMPs, and vesicles (MV). The blot was probed with polyclonal anti-E. coli antibody. Each lane contains 25 μg of total protein. A 30-kDa protein was highly immunoreactive in the vesicle sample. Molecular mass markers (in kilodaltons) are indicated on the left.
FIG. 4
SDS-PAGE profiles of vesicles isolated from various E. coli O157:H7 strains. Each lane contains 15 μg of total protein. Gels were silver stained. Samples were not heat treated prior to loading. Lanes: 1, VDH5; 2, H8302; 3, B19261; 4, DEC3D; 5, ATCC 33694. Molecular mass markers (in kilodaltons) are indicated on the left.
FIG. 5
Agarose gel analysis of PCR products produced with E. coli O157:H7 vesicle-associated DNA. (A) Profiles for stx1 (614 bp) and stx2 (779 bp). Primers for stx1 and stx2 were used with samples in lanes 1 to 5 and 6 to 10, respectively. Samples in each lane were as follows: lanes 1 and 6, intact vesicles; lanes 2 and 7, intact vesicles treated with DNase; lanes 3 and 8, lysed vesicles treated with DNase; lanes 4 and 9, whole cells; and lanes 5 and 10, negative control (PCR cocktail, no template DNA). The lack of fragments in lanes 3 and 8 and the presence of fragments in lanes 2 and 7 indicate that DNA is located in the vesicles. Moreover, the results indicate that DNase treatment was sufficient to digest vesicle-associated DNA. (B) PCR products of eae (863 bp) and uidA (922 bp) primers. Primers for eae and uidA were used with samples in lanes 1 to 3 and lanes 4 to 6, respectively. Lanes: 1 and 4, intact vesicles (treated with DNase); 2 and 5, whole cells; 3 and 6, negative control (PCR cocktail, no template DNA). Molecular size standards (in kilobases) are indicated on the left.
FIG. 6
Immunoblots of Shiga toxin association with E. coli O157:H7 vesicles. The blot was probed with pooled monoclonal antibody against Stx1 and Stx2. Lanes; 1, DEC3D; 2, B4516; 3, H8247; 4, H8302; 5, ATCC 33694; 6, ATCC 43895; 7, B19261; 8, 93-111; 9, DEC8B; 10, VDH5. Note the absence of bands in lane 5 (non-O157, non-Stx-producing isolate) and lane 10 (O157, non-Stx-producing isolate). An arrow indicates the major protein immunologically reactive to pooled monoclonal Stx1 and Stx2 antibody.
FIG. 7
Immunoblot demonstrating that Shiga toxins are located inside vesicles. Intact and lysed vesicles (from E. coli O157:H7) were treated with proteinase K to determine whether Stx is located inside the vesicles. The blot was probed with pooled monoclonal antibody against Stx1 and Stx2. Lanes: 1, intact vesicles treated with proteinase K; 2, lysed vesicles treated with proteinase K; 3, intact vesicles; 4, lysed vesicles. Presence of a band in lane 1 (arrow) and no band in lane 2 indicates that Stx was protected from hydrolysis by virtue of its location within the vesicle (lane 1). Prestained molecular mass markers (in kilodaltons) are indicated on the left.
Similar articles
- Virulence genes of Shiga toxin-producing Escherichia coli isolated from food, animals and humans.
Meng J, Zhao S, Doyle MP. Meng J, et al. Int J Food Microbiol. 1998 Dec 22;45(3):229-35. doi: 10.1016/s0168-1605(98)00163-9. Int J Food Microbiol. 1998. PMID: 9927001 - Vesicle-mediated transfer of virulence genes from Escherichia coli O157:H7 to other enteric bacteria.
Yaron S, Kolling GL, Simon L, Matthews KR. Yaron S, et al. Appl Environ Microbiol. 2000 Oct;66(10):4414-20. doi: 10.1128/AEM.66.10.4414-4420.2000. Appl Environ Microbiol. 2000. PMID: 11010892 Free PMC article. - Semi-automated fluorogenic PCR assays (TaqMan) forrapid detection of Escherichia coli O157:H7 and other shiga toxigenic E. coli.
Sharma VK, Dean-Nystrom EA, Casey TA. Sharma VK, et al. Mol Cell Probes. 1999 Aug;13(4):291-302. doi: 10.1006/mcpr.1999.0251. Mol Cell Probes. 1999. PMID: 10441202 - The locus of enterocyte effacement pathogenicity island of Shiga toxin-producing Escherichia coli O157:H7 and other attaching and effacing E. coli.
Kaper JB. Kaper JB. Jpn J Med Sci Biol. 1998;51 Suppl:S101-7. doi: 10.7883/yoken1952.51.supplement1_s101. Jpn J Med Sci Biol. 1998. PMID: 10211442 Review. No abstract available. - Virulence factors of Escherichia coli O157 and other Shiga toxin-producing E. coli.
Law D. Law D. J Appl Microbiol. 2000 May;88(5):729-45. doi: 10.1046/j.1365-2672.2000.01031.x. J Appl Microbiol. 2000. PMID: 10792533 Review. No abstract available.
Cited by
- A novel pilin subunit from Xenorhabdus nematophila, an insect pathogen, confers pest resistance in tobacco and tomato.
Kumari P, Mahapatro GK, Banerjee N, Sarin NB. Kumari P, et al. Plant Cell Rep. 2015 Nov;34(11):1863-72. doi: 10.1007/s00299-015-1833-6. Epub 2015 Jul 12. Plant Cell Rep. 2015. PMID: 26164296 - Vesiculation from Pseudomonas aeruginosa under SOS.
Maredia R, Devineni N, Lentz P, Dallo SF, Yu J, Guentzel N, Chambers J, Arulanandam B, Haskins WE, Weitao T. Maredia R, et al. ScientificWorldJournal. 2012;2012:402919. doi: 10.1100/2012/402919. Epub 2012 Feb 14. ScientificWorldJournal. 2012. PMID: 22448133 Free PMC article. - Planktonic and Biofilm-Derived Pseudomonas aeruginosa Outer Membrane Vesicles Facilitate Horizontal Gene Transfer of Plasmid DNA.
Johnston EL, Zavan L, Bitto NJ, Petrovski S, Hill AF, Kaparakis-Liaskos M. Johnston EL, et al. Microbiol Spectr. 2023 Mar 22;11(2):e0517922. doi: 10.1128/spectrum.05179-22. Online ahead of print. Microbiol Spectr. 2023. PMID: 36946779 Free PMC article. - Naturally occurring DNA transfer system associated with membrane vesicles in cellulolytic Ruminococcus spp. of ruminal origin.
Klieve AV, Yokoyama MT, Forster RJ, Ouwerkerk D, Bain PA, Mawhinney EL. Klieve AV, et al. Appl Environ Microbiol. 2005 Aug;71(8):4248-53. doi: 10.1128/AEM.71.8.4248-4253.2005. Appl Environ Microbiol. 2005. PMID: 16085810 Free PMC article. - The extracellular vesicle generation paradox: a bacterial point of view.
McMillan HM, Kuehn MJ. McMillan HM, et al. EMBO J. 2021 Nov 2;40(21):e108174. doi: 10.15252/embj.2021108174. Epub 2021 Oct 11. EMBO J. 2021. PMID: 34636061 Free PMC article. Review.
References
- Call J E, Cooke P H, Miller A J. In situ characterization of Clostridium botulinum neurotoxin synthesis and export. J Appl Bacteriol. 1995;79:257–263. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources