Construction and analysis of a library for random insertional mutagenesis in Streptococcus pneumoniae: use for recovery of mutants defective in genetic transformation and for identification of essential genes - PubMed (original) (raw)
Construction and analysis of a library for random insertional mutagenesis in Streptococcus pneumoniae: use for recovery of mutants defective in genetic transformation and for identification of essential genes
M S Lee et al. Appl Environ Microbiol. 1999 May.
Abstract
To explore the use of insertion-duplication mutagenesis (IDM) as a random gene disruption mutagenesis tool for genomic analysis of Streptococcus pneumoniae, a large mutagenic library of chimeric plasmids with 300-bp inserts was constructed. The library was large enough to produce 60,000 independent plasmid clones in Escherichia coli. Sequencing of a random sample of 84 of these clones showed that 85% of the plasmids had inserts which were scattered widely over the genome; 80% of these plasmids had 240- to 360-bp inserts, and 60% of the inserts targeted internal regions of apparent open reading frames. Thus, the library was both complex and highly mutagenic. To evaluate the randomness of mutagenesis during recombination and to test the usefulness of the library for obtaining specific classes of nonessential genes, this library was used to seek competence-related genes by constructing a large pneumococcal transformant library derived from 20,000 mutagenic plasmids. After we screened the mutants exhaustively for transformation defects, 114 competence-related insertion mutations were identified. These competence mutations hit most previously known genes required for transformation as well as a new gene with high similarity to the Bacillus subtilis competence gene comFA. Mapping of the mutation sites at these competence loci showed that the mutagenesis was highly random, with no apparent hot spots. The recovery of a high proportion of competence genes and the absence of hot spots for mutational hits together show that such a transformant library is useful for finding various types of nonessential genes throughout the genome. Since a promoterless lacZ reporter vector was used for the construction of the mutagenic plasmid library, it also serves as a random transcriptional fusion library. Finally, use of a valuable feature of IDM, directed gene targeting, also showed that essential genes, which can be targets for new drug designs, could be identified by simple sequencing and transformation reactions. We estimate that the IDM library used in this study could readily achieve about 90% genome coverage.
Figures
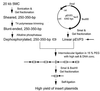
FIG. 1
Strategy for construction of a random mutagenic library of insertion plasmids. Elements of pEVP3 indicated are ori, a replication origin active in E. coli but not in S. pneumoniae; cat, the gene for chloramphenicol acetyltransferase, which confers Cmr to both hosts; and ′lacZ, the promoterless gene for β-galactosidase. Small open boxes, 300-bp pneumococcal fragments; horizontal lines, linear pEVP3 molecules; dots, replication origins; vertical line, vector-vector joint within a concatemer of pEVP3 and inserts. conc., concentrations.
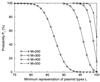
FIG. 2
Dependence of the number of plasmid types (L) represented at least once in a sample on the number of transformants collected (M) after transformation by a pool of 100 plasmids in equimolar concentrations. The probability (PL) of representing at least L plasmid types (Materials and Methods) is shown for several numbers of collected transformants (M).
FIG. 3
Strategy for recovery of sequences adjacent to an integrated pEVP3 plasmid by LMPCR. In the insertion orientation shown, lacZ is transcribed from the same strand as the target gene. Hatched boxes represent duplications of the target region. In this orientation, the targeted gene is truncated at the right end of the left hatched box. Vertical lines indicate the restriction sites closest to the ends of the plasmid insert for each enzyme. Map positions of primers used for LMPCR are shown by flags. Junction fragments produced by digestion of mutant DNA with one of the enzymes shown were attached to a linker oligonucleotide before PCR amplification with primers complementary to the vector and linker. The combinations of restriction enzymes, linkers, PCR primers, and sequencing primers used are shown in Table 2.
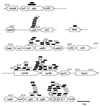
FIG. 4
Distribution of insertions in competence mutants at six loci. Locations, orientations, and structures of plasmid insertions in 114 transformation-defective mutants. Numbers of mutants were 42 at comAB, 38 at cgl, 20 at cel, 4 at cfl, 9 at ccl, and 1 at dal. Mutations are represented above the maps by a pentagon showing the position of the targeting fragment deduced from a sequenced junction. Pentagons pointing to the right indicate vector insertion in the orientation shown in Fig. 3. Open pentagons, insertion-deletions; filled pentagons, insertion-duplications. Putative genes with high levels of similarity to known genes in other organisms are given the prefix “h” (homologue of); those with no significant homology are given “orf” designations. Index genes for these loci are located in TIGR’s data release of 1997 as follows: cclA (contig 4291, bp 1992 to 1333), celA (contig 4139, bp 5654 to 5004), cflA (contig 4155, bp 6454 to 5156), cglA (contig 4194, bp 17495 to 16554), comA (contig 4105, bp 4693 to 6852), and dalA (contig 4179, bp 2168 to 1308). (celB1 and celB2 are fused in strain Rx [35] to form a full-length homologue of B. subtilis comEC.)
Similar articles
- Insertion-duplication mutagenesis in Streptococcus pneumoniae: targeting fragment length is a critical parameter in use as a random insertion tool.
Lee MS, Seok C, Morrison DA. Lee MS, et al. Appl Environ Microbiol. 1998 Dec;64(12):4796-802. doi: 10.1128/AEM.64.12.4796-4802.1998. Appl Environ Microbiol. 1998. PMID: 9835564 Free PMC article. - Isolation and characterization of three Streptococcus pneumoniae transformation-specific loci by use of a lacZ reporter insertion vector.
Pestova EV, Morrison DA. Pestova EV, et al. J Bacteriol. 1998 May;180(10):2701-10. doi: 10.1128/JB.180.10.2701-2710.1998. J Bacteriol. 1998. PMID: 9573156 Free PMC article. - Differential fluorescence induction reveals Streptococcus pneumoniae loci regulated by competence stimulatory peptide.
Bartilson M, Marra A, Christine J, Asundi JS, Schneider WP, Hromockyj AE. Bartilson M, et al. Mol Microbiol. 2001 Jan;39(1):126-35. doi: 10.1046/j.1365-2958.2001.02218.x. Mol Microbiol. 2001. PMID: 11123694 - Genetic transformation in Streptococcus pneumoniae: molecular cloning and characterization of recP, a gene required for genetic recombination.
Rhee DK, Morrison DA. Rhee DK, et al. J Bacteriol. 1988 Feb;170(2):630-7. doi: 10.1128/jb.170.2.630-637.1988. J Bacteriol. 1988. PMID: 2828317 Free PMC article. - Cloning and expression of pneumococcal genes in Streptococcus pneumoniae.
Lacks SA. Lacks SA. Microb Drug Resist. 1997 Winter;3(4):327-37. doi: 10.1089/mdr.1997.3.327. Microb Drug Resist. 1997. PMID: 9442484 Review.
Cited by
- Development of genomic array footprinting for identification of conditionally essential genes in Streptococcus pneumoniae.
Bijlsma JJ, Burghout P, Kloosterman TG, Bootsma HJ, de Jong A, Hermans PW, Kuipers OP. Bijlsma JJ, et al. Appl Environ Microbiol. 2007 Mar;73(5):1514-24. doi: 10.1128/AEM.01900-06. Epub 2007 Jan 19. Appl Environ Microbiol. 2007. PMID: 17261526 Free PMC article. - Exit from competence for genetic transformation in Streptococcus pneumoniae is regulated at multiple levels.
Weng L, Piotrowski A, Morrison DA. Weng L, et al. PLoS One. 2013 May 22;8(5):e64197. doi: 10.1371/journal.pone.0064197. Print 2013. PLoS One. 2013. PMID: 23717566 Free PMC article. - Deciphering the intracellular metabolism of Listeria monocytogenes by mutant screening and modelling.
Schauer K, Geginat G, Liang C, Goebel W, Dandekar T, Fuchs TM. Schauer K, et al. BMC Genomics. 2010 Oct 18;11:573. doi: 10.1186/1471-2164-11-573. BMC Genomics. 2010. PMID: 20955543 Free PMC article. - Identification of a new regulator in Streptococcus pneumoniae linking quorum sensing to competence for genetic transformation.
Lee MS, Morrison DA. Lee MS, et al. J Bacteriol. 1999 Aug;181(16):5004-16. doi: 10.1128/JB.181.16.5004-5016.1999. J Bacteriol. 1999. PMID: 10438773 Free PMC article. - Two distinct functions of ComW in stabilization and activation of the alternative sigma factor ComX in Streptococcus pneumoniae.
Sung CK, Morrison DA. Sung CK, et al. J Bacteriol. 2005 May;187(9):3052-61. doi: 10.1128/JB.187.9.3052-3061.2005. J Bacteriol. 2005. PMID: 15838032 Free PMC article.
References
- Aaberge I S, Eng J, Lermark G, Løvik M. Virulence of Streptococcus pneumoniae in mice: a standardized method for preparation and frozen storage of the experimental bacterial inoculum. Microb Pathog. 1995;18:141–152. - PubMed
- Bult C J, et al. Complete genome sequence of the methanogenic archaeon Methanococcus jannaschii. Science. 1996;273:1058–1073. - PubMed
- Campbell E A, Choi S Y, Masure H R. A competence regulon in Streptococcus pneumoniae revealed by genomic analysis. Mol Microbiol. 1998;27:929–939. - PubMed
- Chen J D, Morrison D A. Cloning of Streptococcus pneumoniae DNA fragments in E. coli requires vectors protected by strong transcriptional terminators. Gene. 1987;55:179–187. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources