Analysis of an even-skipped rescue transgene reveals both composite and discrete neuronal and early blastoderm enhancers, and multi-stripe positioning by gap gene repressor gradients - PubMed (original) (raw)
Analysis of an even-skipped rescue transgene reveals both composite and discrete neuronal and early blastoderm enhancers, and multi-stripe positioning by gap gene repressor gradients
M Fujioka et al. Development. 1999 Jun.
Abstract
The entire functional even-skipped locus of Drosophila melanogaster is contained within a 16 kilobase region. As a transgene, this region is capable of rescuing even-skipped mutant flies to fertile adulthood. Detailed analysis of the 7.7 kb of regulatory DNA 3' of the transcription unit revealed ten novel, independently regulated patterns. Most of these patterns are driven by non-overlapping regulatory elements, including ones for syncytial blastoderm stage stripes 1 and 5, while a single element specifies both stripes 4 and 6. Expression analysis in gap gene mutants showed that stripe 5 is restricted anteriorly by Krüppel and posteriorly by giant, the same repressors that regulate stripe 2. Consistent with the coregulation of stripes 4 and 6 by a single cis-element, both the anterior border of stripe 4 and the posterior border of stripe 6 are set by zygotic hunchback, and the region between the two stripes is 'carved out' by knirps. Thus the boundaries of stripes 4 and 6 are set through negative regulation by the same gap gene domains that regulate stripes 3 and 7 (Small, S., Blair, A. and Levine, M. (1996) Dev. Biol. 175, 314-24), but at different concentrations. The 3' region also contains a single element for neurogenic expression in ganglion mother cells 4-2a and 1-1a, and neurons derived from them (RP2, a/pCC), suggesting common regulators in these lineages. In contrast, separable elements were found for expression in EL neurons, U/CQ neurons and the mesoderm. The even-skipped 3' untranslated region is required to maintain late stage protein expression in RP2 and a/pCC neurons, and appears to affect protein levels rather than mRNA levels. Additionally, a strong pairing-sensitive repression element was localized to the 3' end of the locus, but was not found to contribute to efficient functional rescue.
Figures
Fig. 1
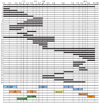
A deletion analysis of the eve 3′ region. Numbers along the top show distance (in kb) to the transcription start site of the eve locus. Regions of the sequence included in each construct are indicated by thick lines, and deleted regions by thin lines. All fragments were placed downstream of an eve promoter-lacZ reporter in a pCaSpeR-based vector (see Materials and Methods). Five to ten independent transgenic lines were analyzed for each construct. Boxes at the bottom indicate minimal elements required to drive the following lacZ expression: _ftz_-like, 7 stripes in the even-numbered parasegments; EL, EL neurons in the central nervous system; CQ, GMC 7-1a and CQ neurons; early APR, early anal plate ring; late APR, late anal plate ring; 4/6, blastoderm stripes 4 and 6; muscle, mesodermal precursors; 1, blastoderm stripe 1; 5, blastoderm stripe 5; RP2,a/pCC, GMCs 4-2a and 1-1a, and neurons RP2, aCC and pCC. PSR indicates the region causing pairing-enhanced repression of the mini-white gene.
Fig. 2
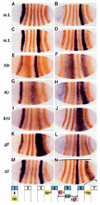
Elements for early stripes 1, 5 and 4+6 are negatively regulated by gap genes. In situ hybridization was performed for lacZ mRNA (blue), followed by staining with anti-Eve antibody (orange). (A) lacZ expression driven by the region +6.6 to +7.4 kb; (B) by the region +7.4 to +8.4 kb; (C) by the region +6.6 to +8.4 kb; (D) by the region +4.0 to 5.2 kb. The transgenic lines in C and D were crossed into several gap gene mutant backgrounds and stained as above. Staining patterns in these mutant embryos are shown below the corresponding wild-type pattern: hb (E,F), Kr (G,H), kni (I,J), gt (K,L) and tll (M,N). A summary of the apparent gap gene regulatory influences on stripes 1, 4, 5 and 6 is diagrammed at the bottom.
Fig. 3
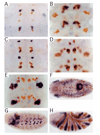
Isolated elements drive various components of tissue-specific expression. In situ hybridization was performed for lacZ mRNA (blue), followed by staining with anti-Eve antibody (orange). (A) lacZ expression in GMC 7-1a progeny at early stage 11 driven by the region + 3.5 to +4.3 kb. (B) lacZ expression in CQ neurons at stage 15 in the same transgenic line as in A. (C) lacZ expression in neurons RP2, aCC, and pCC at stage 11 driven by the region +7.9 to +9.2 kb. Note that at this stage lacZ expression is strong. (D) lacZ expression in RP2, aCC and pCC neurons at stage 15 in the same transgenic line as in C. Overall expression of lacZ at this stage is faint. (E) lacZ expression in EL neurons at stage 15 driven by the region +1.9 to +3.2 kb. (F) lacZ expression in the anal plate ring at stage 12 driven by the region +2.6 to +4.8 kb. (G) lacZ expression in muscle precursor cells at stage 11 driven by the fragment +5.8 to +6.6 kb. (H) lacZ expression in the anterior portion of even-numbered parasegments driven by the region +1.5 to +2.6 kb.
Fig. 4
The eve 3′ UTR is required for high level protein expression in RP2 and aCC/pCC neurons at later stages. (A-D) Wild type; (E-H) transgenic line carrying lacZ construct driven by the +7.9 to + 9.2 kb fragment, with the α-tubulin 3′ UTR (See Materials and Methods); diffuse ectopic expression due to a position effect is visible. (I-L) Transgenic line carrying lacZ construct driven by the same +7.9 to + 9.2 kb fragment, but with the eve 3′ UTR; again, there is diffuse ectopic expression, particularly at stage 11. (A,C,E,G,I,K) mRNA expression visualized by in situ hybridization with appropriate probe; (B,D,F,H,J,L) protein expression visualized by staining with appropriate antibodies. (A,C) eve mRNA; (B,D) Eve protein; (E,G,I,K) lacZ mRNA; (F,H,J,L) β-galactosidase protein. Expression in RP2, aCC and pCC neurons, at stage 11 (E,F,I,J); at stage 15 (G,H,K,L). Arrows, a representative neuron in a representative line, showing increased protein staining at stage 15 with the eve 3′ UTR (in L) relative to that with the α-tubulin 3′ UTR (in H).
Similar articles
- The even-skipped locus is contained in a 16-kb chromatin domain.
Sackerson C, Fujioka M, Goto T. Sackerson C, et al. Dev Biol. 1999 Jul 1;211(1):39-52. doi: 10.1006/dbio.1999.9301. Dev Biol. 1999. PMID: 10373303 - Regulation of two pair-rule stripes by a single enhancer in the Drosophila embryo.
Small S, Blair A, Levine M. Small S, et al. Dev Biol. 1996 May 1;175(2):314-24. doi: 10.1006/dbio.1996.0117. Dev Biol. 1996. PMID: 8626035 - Functional analysis of eve stripe 2 enhancer evolution in Drosophila: rules governing conservation and change.
Ludwig MZ, Patel NH, Kreitman M. Ludwig MZ, et al. Development. 1998 Mar;125(5):949-58. doi: 10.1242/dev.125.5.949. Development. 1998. PMID: 9449677 - The repressor activity of Even-skipped is highly conserved, and is sufficient to activate engrailed and to regulate both the spacing and stability of parasegment boundaries.
Fujioka M, Yusibova GL, Patel NH, Brown SJ, Jaynes JB. Fujioka M, et al. Development. 2002 Oct;129(19):4411-21. doi: 10.1242/dev.129.19.4411. Development. 2002. PMID: 12223400 Free PMC article. - From gradients to stripes in Drosophila embryogenesis: filling in the gaps.
Rivera-Pomar R, Jäckle H. Rivera-Pomar R, et al. Trends Genet. 1996 Nov;12(11):478-83. doi: 10.1016/0168-9525(96)10044-5. Trends Genet. 1996. PMID: 8973159 Review.
Cited by
- Microtubules are organized independently of the centrosome in Drosophila neurons.
Nguyen MM, Stone MC, Rolls MM. Nguyen MM, et al. Neural Dev. 2011 Dec 6;6:38. doi: 10.1186/1749-8104-6-38. Neural Dev. 2011. PMID: 22145670 Free PMC article. - Postsynaptic protein kinase A reduces neuronal excitability in response to increased synaptic excitation in the Drosophila CNS.
Baines RA. Baines RA. J Neurosci. 2003 Sep 24;23(25):8664-72. doi: 10.1523/JNEUROSCI.23-25-08664.2003. J Neurosci. 2003. PMID: 14507965 Free PMC article. - Multiple enhancers ensure precision of gap gene-expression patterns in the Drosophila embryo.
Perry MW, Boettiger AN, Levine M. Perry MW, et al. Proc Natl Acad Sci U S A. 2011 Aug 16;108(33):13570-5. doi: 10.1073/pnas.1109873108. Epub 2011 Aug 8. Proc Natl Acad Sci U S A. 2011. PMID: 21825127 Free PMC article. - Ceramides And Stress Signalling Intersect With Autophagic Defects In Neurodegenerative Drosophila blue cheese (bchs) Mutants.
Hebbar S, Sahoo I, Matysik A, Argudo Garcia I, Osborne KA, Papan C, Torta F, Narayanaswamy P, Fun XH, Wenk MR, Shevchenko A, Schwudke D, Kraut R. Hebbar S, et al. Sci Rep. 2015 Dec 7;5:15926. doi: 10.1038/srep15926. Sci Rep. 2015. PMID: 26639035 Free PMC article. - Sex-specific pattern formation during early Drosophila development.
Manu, Ludwig MZ, Kreitman M. Manu, et al. Genetics. 2013 May;194(1):163-73. doi: 10.1534/genetics.112.148205. Epub 2013 Feb 14. Genetics. 2013. PMID: 23410834 Free PMC article.
References
- Ahringer J. Posterior patterning by the Caenorhabditis elegans even-skipped homolog vab-7. Genes Dev. 1996;10:1120–1130. - PubMed
- Bate M. The mesoderm and its derivatives. In: Bate M, Martinez Arias A, editors. The development of Drosophila melanogaster. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 1993. pp. 1013–1090.
- Bodmer R. The gene tinman is required for specification of the heart and visceral muscles in Drosophila. Development. 1993;118:719–729. - PubMed
- Bossing T, Udolph G, Doe CQ, Technau GM. The embryonic central nervous system lineages of Drosophila melanogaster. I. Neuroblast lineages derived from the ventral half of the neuroectoderm. Dev. Biol. 1996;179:41–64. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous