Characterization of differentially expressed genes in purified Drosophila follicle cells: toward a general strategy for cell type-specific developmental analysis - PubMed (original) (raw)
Characterization of differentially expressed genes in purified Drosophila follicle cells: toward a general strategy for cell type-specific developmental analysis
Z Bryant et al. Proc Natl Acad Sci U S A. 1999.
Abstract
Axis formation in Drosophila depends on correct patterning of the follicular epithelium and on signaling between the germ line and soma during oogenesis. We describe a method for identifying genes expressed in the follicle cells with potential roles in axis formation. Follicle cells are purified from whole ovaries by enzymatic digestion, filtration, and fluorescence-activated cell sorting (FACS). Two strategies are used to obtain complementary cell groups. In the first strategy, spatially restricted subpopulations are marked for FACS selection using a green fluorescent protein (GFP) reporter. In the second, cells are purified from animals mutant for the epidermal growth factor receptor ligand gurken (grk) and from their wild-type siblings. cDNA from these samples of spatially restricted or genetically mutant follicle cells is used in differential expression screens employing PCR-based differential display or hybridization to a cDNA microarray. Positives are confirmed by in situ hybridization to whole mounts. These methods are found to be capable of identifying both spatially restricted and grk-dependent transcripts. Results from our pilot screens include (i) the identification of a homologue of the immunophilin FKBP-12 with dorsal anterior expression in egg chambers, (ii) the discovery that the ecdysone-inducible nuclear hormone receptor gene E78 is regulated by grk during oogenesis and is required for proper dorsal appendage formation, and (iii) the identification of a Drosophila homologue of the human SET-binding factor gene SBF1 with elevated transcription in grk mutant egg chambers.
Figures
Figure 1
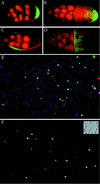
Marking and purification of follicle cell subgroups. (A–D) GAL4 expression patterns in egg chambers. Posterior is to the right. Nuclei are stained with propidium iodide, shown as red. Green fluorescence reflects GAL4-activated transcription of GFP. (A and B) A62 GAL4/UAS-GFP. (A) Stage 7 egg chamber showing GFP expression in posterior follicle cells. (B) Stage 10 egg chamber showing GFP expression in posterior follicle cells and in border cells (small anterior group). (C and D) 55B GAL4/UAS-GFP. Expression is seen in lateral follicle cells in stage 8 (C) through stage 10 (D) egg chambers. (E and F) Fluorescence images of cells released from A62 GAL4/UAS-GFP ovaries, before (E) and after (F) purification of GFP-positive cells by FACS. All nuclei stain blue with Hoechst 33342; the nuclei of dead cells stain red with propidium iodide (cells were restained after sorting). GFP fluorescence is shown in green. (Inset) Nomarski image of A62− cells after sorting.
Figure 2
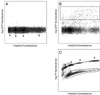
FACS analysis of follicle cells. Cells are plotted as fluorescence events on the axes shown. (A) Wild-type follicle cells. There are very few events in the region of high GFP fluorescence. Arrows indicate concentrated regions along the Hoechst axis roughly corresponding to the four major ploidies in the population. (B) A62 GAL4/UAS-GFP. A population of GFP-positive cells can clearly be distinguished. The rectangle represents the approximate sort window used to select GFP-positive cells. (C) Live/dead selection. The approximate sort window used to select live cells is overlaid on the plot. The upper streak, which is high in propidium iodide fluorescence, represents dead cells, which are excluded from the sorted population. The separation of cells of different ploidies can most easily be seen in the dead cells (arrows), whose exposed nuclei can readily equilibrate with the Hoechst dye.
Figure 3
Autoradiographs showing that GFP and pointed P1 are enriched in the GFP-selected, posterior-derived A62+ cells. Amplified cDNA was prepared from sorted A62+ and A62− cells and blotted onto a nylon membrane. This membrane was hybridized to three successive radioactive oligonucleotides: the universally expressed ribosomal protein rp49, the reporter transgene GFP, and the posterior-expressed transcript pointed P1.
Figure 4
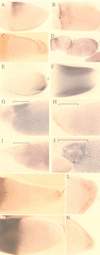
In situ hybridization patterns of microarray (A–D) and differential display positives (E–L). Egg chambers are shown with posterior to the right. (A–C) GM07659, a gene with homology to human FKBP-12. (A) Lateral view of a stage 10A egg chamber, showing expression restricted to a dorsal anterior triangle of follicle cells. (B) Dorsal view of a stage 10B egg chamber, showing dorsal anterior staining. (C) No staining is seen in _grk_2B6/_grk_DC9 egg chambers. (D) GM04985, a splice variant of the E75 nuclear hormone receptor gene, hybridizes to follicle cells over the posterior (arrows) of stage 6–9 egg chambers. (E and F) The vitelline membrane protein VM34C. Stage 8 (E) and stage 10 (F) egg chambers, showing expression in the columnar follicle cells over the oocyte. Staining is excluded from the posterior (arrows). (G–J) The The ecdysone-responsive nuclear hormone receptor E78. (G) Hybridization of an RNA antisense probe to a stage 12 egg chamber, showing expression in the dorsal anterior follicle cells (bracket). No staining in the follicle cells is seen with an RNA sense probe (H) nor in grk egg chambers (I). (J) A stage 14 egg chamber showing expression in the follicle cells covering the dorsal appendages (bracket). (K–N) Drosophila SET-binding factor 1. In _grk_2B6/_grk_DC9 flies, staining can be detected in follicle cells over the oocytes of stage 8 through stage 11 egg chambers (K), with the highest concentration in the posterior (arrow). (L) Close-up of staining in the posterior follicle cells (stage 10). (M) In the wild-type egg chambers of grk/CyO flies, hybridization is faint or undetectable in the follicle cells surrounding the oocyte, although staining is evident in the nurse cells. (N) Stage 9 _grk_2B6/_grk_DC9 egg chamber
Figure 5
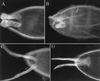
(A–C) Chorions of eggs laid by E78 mutant flies. Dorsal side is facing the camera; anterior is to the left. Twenty percent of eggshells have broadened and/or shortened dorsal appendages (A and B); the remainder have wild-type appearance (C). (D) Eggs laid by hs-E78B flies have fused appendages.
Similar articles
- Post-transcriptional regulation of gurken by encore is required for axis determination in Drosophila.
Hawkins NC, Van Buskirk C, Grossniklaus U, Schüpbach T. Hawkins NC, et al. Development. 1997 Dec;124(23):4801-10. doi: 10.1242/dev.124.23.4801. Development. 1997. PMID: 9428416 - A novel follicle-cell-dependent dominant female sterile allele, StarKojak, alters receptor tyrosine kinase signaling in Drosophila.
Ruden DM, Wang X, Cui W, Mori D, Alterman M. Ruden DM, et al. Dev Biol. 1999 Mar 15;207(2):393-407. doi: 10.1006/dbio.1998.9148. Dev Biol. 1999. PMID: 10068471 - Combined activities of Gurken and decapentaplegic specify dorsal chorion structures of the Drosophila egg.
Peri F, Roth S. Peri F, et al. Development. 2000 Feb;127(4):841-50. doi: 10.1242/dev.127.4.841. Development. 2000. PMID: 10648242 - Signaling pathways that establish the dorsal-ventral pattern of the Drosophila embryo.
Morisato D, Anderson KV. Morisato D, et al. Annu Rev Genet. 1995;29:371-99. doi: 10.1146/annurev.ge.29.120195.002103. Annu Rev Genet. 1995. PMID: 8825480 Review. - RNA localization and translational regulation during axis specification in the Drosophila oocyte.
Cooperstock RL, Lipshitz HD. Cooperstock RL, et al. Int Rev Cytol. 2001;203:541-66. doi: 10.1016/s0074-7696(01)03016-9. Int Rev Cytol. 2001. PMID: 11131526 Review.
Cited by
- Have microarrays failed to deliver for developmental biology?
Livesey R. Livesey R. Genome Biol. 2002 Aug 27;3(9):comment2009. doi: 10.1186/gb-2002-3-9-comment2009. Epub 2002 Aug 27. Genome Biol. 2002. PMID: 12225576 Free PMC article. - Identification and comparative analysis of the peptidyl-prolyl cis/trans isomerase repertoires of H. sapiens, D. melanogaster, C. elegans, S. cerevisiae and Sz. pombe.
Pemberton TJ, Kay JE. Pemberton TJ, et al. Comp Funct Genomics. 2005;6(5-6):277-300. doi: 10.1002/cfg.482. Comp Funct Genomics. 2005. PMID: 18629211 Free PMC article. - Analysis of Cell Cycle Switches in Drosophila Oogenesis.
Jia D, Huang YC, Deng WM. Jia D, et al. Methods Mol Biol. 2015;1328:207-16. doi: 10.1007/978-1-4939-2851-4_15. Methods Mol Biol. 2015. PMID: 26324440 Free PMC article. - Following the 'tracks': Tramtrack69 regulates epithelial tube expansion in the Drosophila ovary through Paxillin, Dynamin, and the homeobox protein Mirror.
Peters NC, Thayer NH, Kerr SA, Tompa M, Berg CA. Peters NC, et al. Dev Biol. 2013 Jun 15;378(2):154-69. doi: 10.1016/j.ydbio.2013.03.017. Epub 2013 Mar 30. Dev Biol. 2013. PMID: 23545328 Free PMC article. - Purification of Low-abundant Cells in the Drosophila Visual System.
Peng J, Santiago IJ, Pecot MY. Peng J, et al. J Vis Exp. 2018 Sep 26;(139):58474. doi: 10.3791/58474. J Vis Exp. 2018. PMID: 30320761 Free PMC article.
References
- Liang P, Pardee A B. Science. 1992;257:967–971. - PubMed
- Liang P, Bauer D, Averboukh L, Warthoe P, Rohrwild M, Muller H, Strauss M, Pardee A B. Methods Enzymol. 1995;254:304–321. - PubMed
- Schena M, Shalon D, Davis R W, Brown P O. Science. 1995;270:467–470. - PubMed
- Ramsay G. Nat Biotechnol. 1998;16:40–44. - PubMed
- Krasnow M A, Cumberledge S, Manning G, Herzenberg L A, Nolan G P. Science. 1991;251:81–85. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials