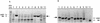Efficient approach to unique single-nucleotide polymorphism discovery - PubMed (original) (raw)
Efficient approach to unique single-nucleotide polymorphism discovery
P Taillon-Miller et al. Genome Res. 1999 May.
Abstract
Single-nucleotide polymorphisms (SNPs) are the most frequently found DNA sequence variations in the human genome. It has been argued that a dense set of SNP markers can be used to identify genetic factors associated with complex disease traits. Because all high-throughput genotyping methods require precise sequence knowledge of the SNPs, any SNP discovery approach must involve both the determination of DNA sequence and allele frequencies. Furthermore, high-throughput genotyping also requires a genomic DNA amplification step, making it necessary to develop sequence-tagged sites (STSs) that amplify only the DNA fragment containing the SNP and nothing else from the rest of the genome. In this report, we demonstrate the utility of a SNP-screening approach that yields the DNA sequence and allele frequency information while screening out duplications with minimal cost and effort. Our approach is based on the use of a homozygous complete hydatidiform mole (CHM) as the reference. With this homozygous reference, one can identify and estimate the allele frequencies of common SNPs with a pooled DNA-sequencing approach (rather than having to sequence numerous individuals as is commonly done). More importantly, the CHM reference is preferable to a single individual reference because it reveals readily any duplicated regions of the genome amplified by the PCR assay before the duplicated sequences are found in GenBank. This approach reduces the cost of SNP discovery by 60% and eliminates the costly development of SNP markers that cannot be amplified uniquely from the genome.
Figures
Figure 1
The results of scanning sWXD3868 for SNPs by method 1 is shown in A. CEPH parents are 1, 2, 3, 4, and the CEPH population pool is 5 (additional detail about DNAs used are included in Methods). Sequencing was done with the dRhodamine terminators in A. The results for method 2 are shown in B. The CHM1 is sample 6 and the CEPH population pool is sample 5. Sequencing was done with the BigDye terminators in B. (↓) SNP locations. The small blue underhand for the T peak (↓) in B, sample 6 is a common sequencing artifact and was seen in a number of T peaks in this sequencing trace.
Figure 2
The results of scanning sWXD3654 for SNPs by methods 1 and 2 are shown. (Method 1) CEPH parents are 1, 2, 3, and 4 and the CEPH population pool is 5 (additional detail about DNAs used is included in Methods); (method 2) CHM1 is sample 6, and the CEPH population pool is sample 5. The CEPH population pool is shown only once. Sequencing was done with the BigDye terminators. (↓) SNP locations.
Figure 3
Results of testing STSs sWXD3555 (A) and sWXD3857 (B) against the chromosome panel pools (1–8), human genomic DNA (9), hamster genomic DNA (10), and mouse genomic DNA (11). sWXD3555 amplified only pools 2 and 8 and the human control, indicating that it was found on the X chromosome. sWXD3857 amplified all eight pools and the human control, indicating that it was found on multiple chromosomes. (Shaded arrows) The 700- and 1000-bp molecular weight standards (50–2000 bp, Bio-Rad, Hercules, CA); (black arrows) the specific PCR product. Detail descriptions of the chromosome panel pools are included in Methods.
Similar articles
- SNP-PHAGE--High throughput SNP discovery pipeline.
Matukumalli LK, Grefenstette JJ, Hyten DL, Choi IY, Cregan PB, Van Tassell CP. Matukumalli LK, et al. BMC Bioinformatics. 2006 Oct 23;7:468. doi: 10.1186/1471-2105-7-468. BMC Bioinformatics. 2006. PMID: 17059604 Free PMC article. - SNP discovery and allele frequency estimation by deep sequencing of reduced representation libraries.
Van Tassell CP, Smith TP, Matukumalli LK, Taylor JF, Schnabel RD, Lawley CT, Haudenschild CD, Moore SS, Warren WC, Sonstegard TS. Van Tassell CP, et al. Nat Methods. 2008 Mar;5(3):247-52. doi: 10.1038/nmeth.1185. Epub 2008 Feb 24. Nat Methods. 2008. PMID: 18297082 - Paternal origins of complete hydatidiform moles proven by whole genome single-nucleotide polymorphism haplotyping.
Fan JB, Surti U, Taillon-Miller P, Hsie L, Kennedy GC, Hoffner L, Ryder T, Mutch DG, Kwok PY. Fan JB, et al. Genomics. 2002 Jan;79(1):58-62. doi: 10.1006/geno.2001.6676. Genomics. 2002. PMID: 11827458 - SNP genotyping: technologies and biomedical applications.
Kim S, Misra A. Kim S, et al. Annu Rev Biomed Eng. 2007;9:289-320. doi: 10.1146/annurev.bioeng.9.060906.152037. Annu Rev Biomed Eng. 2007. PMID: 17391067 Review. - Detection of single nucleotide polymorphisms.
Kwok PY, Chen X. Kwok PY, et al. Curr Issues Mol Biol. 2003 Apr;5(2):43-60. Curr Issues Mol Biol. 2003. PMID: 12793528 Review.
Cited by
- Single-nucleotide polymorphisms in soybean.
Zhu YL, Song QJ, Hyten DL, Van Tassell CP, Matukumalli LK, Grimm DR, Hyatt SM, Fickus EW, Young ND, Cregan PB. Zhu YL, et al. Genetics. 2003 Mar;163(3):1123-34. doi: 10.1093/genetics/163.3.1123. Genetics. 2003. PMID: 12663549 Free PMC article. - A SNP resource for human chromosome 22: extracting dense clusters of SNPs from the genomic sequence.
Dawson E, Chen Y, Hunt S, Smink LJ, Hunt A, Rice K, Livingston S, Bumpstead S, Bruskiewich R, Sham P, Ganske R, Adams M, Kawasaki K, Shimizu N, Minoshima S, Roe B, Bentley D, Dunham I. Dawson E, et al. Genome Res. 2001 Jan;11(1):170-8. doi: 10.1101/gr.156901. Genome Res. 2001. PMID: 11156626 Free PMC article. - Ordered catenation of sequence-tagged sites and multiplexed SNP genotyping by sequencing.
Higasa K, Hayashi K. Higasa K, et al. Nucleic Acids Res. 2002 Feb 1;30(3):E11. doi: 10.1093/nar/30.3.e11. Nucleic Acids Res. 2002. PMID: 11809899 Free PMC article. - An initial map of insertion and deletion (INDEL) variation in the human genome.
Mills RE, Luttig CT, Larkins CE, Beauchamp A, Tsui C, Pittard WS, Devine SE. Mills RE, et al. Genome Res. 2006 Sep;16(9):1182-90. doi: 10.1101/gr.4565806. Epub 2006 Aug 10. Genome Res. 2006. PMID: 16902084 Free PMC article. - Estimating Copy-Number Proportions: The Comeback of Sanger Sequencing.
Seroussi E. Seroussi E. Genes (Basel). 2021 Feb 17;12(2):283. doi: 10.3390/genes12020283. Genes (Basel). 2021. PMID: 33671263 Free PMC article.
References
- Collins FS, Guyer MS, Chakravarti A. Variations on a theme: Cataloging human DNA sequence variation. Science. 1997;278:1580–1581. - PubMed
- Dausset J, Cann H, Cohen D, Lathrop M, Lalouel JM, White R. Centre d’etude du polymorphisme humain (CEPH): Collaborative genetic mapping of the human genome. Genomics. 1990;6:575–577. - PubMed
- Eichler EE, Lu F, Shen Y, Antonacci R, Jurecic V, Doggett NA, Moyzis RK, Baldini A, Gibbs RA, Nelson DL. Duplication of a gene-rich cluster between 16p11.1 and Xq28: A novel pericentromeric-directed mechanism for paralogous genome evolution. Hum Mol Genet. 1996;5:899–912. - PubMed
- Eichler EE, Budarf ML, Rocchi M, Deaven LL, Doggett NA, Baldini A, Nelson DL, Mohrenweiser HW. Interchromosomal duplications of the adrenoleukodystrophy locus: A phenomenon of pericentromeric plasticity. Hum Mol Genet. 1997;6:991–1002. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials