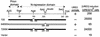Two prion-inducing regions of Ure2p are nonoverlapping - PubMed (original) (raw)
Two prion-inducing regions of Ure2p are nonoverlapping
M L Maddelein et al. Mol Cell Biol. 1999 Jun.
Abstract
Ure2p of Saccharomyces cerevisiae normally functions in blocking utilization of a poor nitrogen source when a good nitrogen source is available. The non-Mendelian genetic element [URE3] is a prion (infectious protein) form of Ure2p, so that overexpression of Ure2p induces the de novo appearance of infectious [URE3]. Earlier studies defined a prion domain comprising Ure2p residues 1 to 64 and a nitrogen regulation domain included in residues 66 to 354. We find that deletion of individual runs of asparagine within the prion domain reduce prion-inducing activity. Although residues 1 to 64 are sufficient for prion induction, the fragment from residues 1 to 80 is a more efficient inducer of [URE3]. In-frame deletion of a region around residue 224 does not affect nitrogen regulation but does eliminate prion induction by the remainder of Ure2p. Larger deletions removing the region around residue 224 and more of the C-terminal part of Ure2p restore prion-inducing ability. A fragment of Ure2p lacking the original prion domain does not induce [URE3], but surprisingly, further deletion of residues 151 to 157 and 348 to 354 leaves a fragment that can do so. The region from 66 to 80 and the region around residue 224 are both necessary for this second prion-inducing activity. Thus, each of two nonoverlapping parts of Ure2p is sufficient to induce the appearance of the [URE3] prion.
Figures
FIG. 1
Asparagine runs are important for prion-inducing activity of Ure2p. A diploid strain (3469) was transformed with each plasmid and grown on galactose for 3 days to overproduce the indicated part of Ure2p; cells were plated on SD with ureidosuccinate in place of uracil, selective for [URE3] cells. ■, active in prion induction; □, inactive in prion induction. The top line shows low prion induction activity. The N-terminal part of the Ure2p sequence is shown below, with the regions deleted in p773, p774, p776, and p778 underlined. comple, complementation.
FIG. 2
Residues 66 to 80 enhance prion induction. ■, active in prion induction; □, inactive in prion induction. The top line shows low prion induction activity. comple, complementation.
FIG. 3
Ure2p residues 221 to 227 are needed for optimal [URE3] induction by intact Ure2p. ■, active in prion induction; □, inactive in prion induction. The top line shows low prion induction activity. comple, complementation.
FIG. 4
A fragment of Ure2p lacking the 1–64 prion domain induces [URE3]. ■, active in prion induction; □, inactive in prion induction. The top line shows low prion induction activity. comple, complementation.
FIG. 5
Asparagine runs are important but not essential for prion induction by derepressed Ure2p fragments. Note that C3, B2, and A2 all have identical C-terminal tails. The C-terminal tail of 679 differs from those of P215 and P408. ■, active in prion induction; □, inactive in prion induction. The top line shows low prion induction activity. comple, complementation.
FIG. 6
C-terminal tails can affect prion induction efficiency. ■, active in prion induction; □, inactive in prion induction. The top line shows low prion induction activity. comple, complementation.
FIG. 7
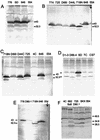
Steady-state levels of mutant Ure2 proteins measured by Western blotting. Construct names are as in Table 1 and Fig. 1 to 6. 554 is the vector alone, and 646 expresses full-length Ure2p. Anti-Ure2p antibodies used were U2 and affinity-purified K1, K2, K3, and C3 (A), affinity-purified U2 (B), U2, K2, and C3 (C and D), U2 (E), and affinity-purified U2, K1, K2, K3, and C3 (F). SD is 646SD. Positions of selected size markers are shown in kilodaltons. Equal amounts of total protein were analyzed.
FIG. 8
Prion-promoting and -inhibiting domains of Ure2p compared to the glutathione _S_-transferase homology region and the nitrogen regulation domain.
Similar articles
- Prion-inducing domain of yeast Ure2p and protease resistance of Ure2p in prion-containing cells.
Masison DC, Wickner RB. Masison DC, et al. Science. 1995 Oct 6;270(5233):93-5. doi: 10.1126/science.270.5233.93. Science. 1995. PMID: 7569955 - The [URE3] prion is an aggregated form of Ure2p that can be cured by overexpression of Ure2p fragments.
Edskes HK, Gray VT, Wickner RB. Edskes HK, et al. Proc Natl Acad Sci U S A. 1999 Feb 16;96(4):1498-503. doi: 10.1073/pnas.96.4.1498. Proc Natl Acad Sci U S A. 1999. PMID: 9990052 Free PMC article. - The prion model for [URE3] of yeast: spontaneous generation and requirements for propagation.
Masison DC, Maddelein ML, Wickner RB. Masison DC, et al. Proc Natl Acad Sci U S A. 1997 Nov 11;94(23):12503-8. doi: 10.1073/pnas.94.23.12503. Proc Natl Acad Sci U S A. 1997. PMID: 9356479 Free PMC article. - Prions in Saccharomyces and Podospora spp.: protein-based inheritance.
Wickner RB, Taylor KL, Edskes HK, Maddelein ML, Moriyama H, Roberts BT. Wickner RB, et al. Microbiol Mol Biol Rev. 1999 Dec;63(4):844-61, table of contents. doi: 10.1128/MMBR.63.4.844-861.1999. Microbiol Mol Biol Rev. 1999. PMID: 10585968 Free PMC article. Review. - Prions of yeast as heritable amyloidoses.
Wickner RB, Taylor KL, Edskes HK, Maddelein ML, Moriyama H, Roberts BT. Wickner RB, et al. J Struct Biol. 2000 Jun;130(2-3):310-22. doi: 10.1006/jsbi.2000.4250. J Struct Biol. 2000. PMID: 10940235 Review.
Cited by
- Interactions among prions and prion "strains" in yeast.
Bradley ME, Edskes HK, Hong JY, Wickner RB, Liebman SW. Bradley ME, et al. Proc Natl Acad Sci U S A. 2002 Dec 10;99 Suppl 4(Suppl 4):16392-9. doi: 10.1073/pnas.152330699. Epub 2002 Jul 30. Proc Natl Acad Sci U S A. 2002. PMID: 12149514 Free PMC article. - A promiscuous prion: efficient induction of [URE3] prion formation by heterologous prion domains.
Ross CD, McCarty BR, Hamilton M, Ben-Hur A, Ross ED. Ross CD, et al. Genetics. 2009 Nov;183(3):929-40. doi: 10.1534/genetics.109.109322. Epub 2009 Sep 14. Genetics. 2009. PMID: 19752212 Free PMC article. - The mechanism of prion inhibition by HET-S.
Greenwald J, Buhtz C, Ritter C, Kwiatkowski W, Choe S, Maddelein ML, Ness F, Cescau S, Soragni A, Leitz D, Saupe SJ, Riek R. Greenwald J, et al. Mol Cell. 2010 Jun 25;38(6):889-99. doi: 10.1016/j.molcel.2010.05.019. Mol Cell. 2010. PMID: 20620958 Free PMC article. - PrionHome: a database of prions and other sequences relevant to prion phenomena.
Harbi D, Parthiban M, Gendoo DM, Ehsani S, Kumar M, Schmitt-Ulms G, Sowdhamini R, Harrison PM. Harbi D, et al. PLoS One. 2012;7(2):e31785. doi: 10.1371/journal.pone.0031785. Epub 2012 Feb 20. PLoS One. 2012. PMID: 22363733 Free PMC article. - Potential roles for prions and protein-only inheritance in cancer.
Antony H, Wiegmans AP, Wei MQ, Chernoff YO, Khanna KK, Munn AL. Antony H, et al. Cancer Metastasis Rev. 2012 Jun;31(1-2):1-19. doi: 10.1007/s10555-011-9325-9. Cancer Metastasis Rev. 2012. PMID: 22138778 Free PMC article. Review.
References
- Alper T, Cramp W A, Haig D A, Clarke M C. Does the agent of scrapie replicate without nucleic acid? Nature. 1967;214:764–766. - PubMed
- Bolton D C, McKinley M P, Prusiner S B. Identification of a protein that purifies with the scrapie prion. Science. 1982;218:1309–1311. - PubMed
- Bruce M E, McConnell I, Fraser H, Dickinson A G. The disease characteristics of different strains of scrapie in Sinc congenic mouse lines: implications for the nature of the agent and host control of pathogenesis. J Gen Virol. 1991;72:595–603. - PubMed
- Bueler H, Aguzzi A, Sailer A, Greiner R-A, Autenried P, Aguet M, Weissmann C. Mice devoid of PrP are resistant to scrapie. Cell. 1993;73:1339–1347. - PubMed
- Carlson G A, Kingsbury D T, Goodman P A, Coleman S, Marshall S T, DeArmond S, Westaway D, Prusiner S B. Linkagae of prion protein and scrapie incubation time genes. Cell. 1986;46:503–511. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases