Interruption of multiple cellular processes in HT-29 epithelial cells by Pseudomonas aeruginosa exoenzyme S - PubMed (original) (raw)
Interruption of multiple cellular processes in HT-29 epithelial cells by Pseudomonas aeruginosa exoenzyme S
J C Olson et al. Infect Immun. 1999 Jun.
Abstract
Exoenzyme S (ExoS), an ADP-ribosylating enzyme produced by the opportunistic pathogen Pseudomonas aeruginosa, is directly translocated into eukaryotic cells by bacterial contact. Within the cell, ExoS ADP-ribosylates the cell signaling protein Ras and causes inhibition of DNA synthesis and alterations in cytoskeletal structure. To further understand the interrelationship of the different cellular effects of ExoS, functional analyses were performed on HT-29 epithelial cells after exposure to ExoS-producing P. aeruginosa 388 and the non-ExoS-producing strain 388DeltaS. Two different mechanisms of morphological alteration were identified: (i) a more-transient and less-severe cell rounding caused by the non-ExoS-producing strain 388DeltaS and (ii) a more-severe, long-term cell rounding caused by ExoS-producing strain 388. Long-term effects of ExoS on cell morphology occurred in conjunction with ExoS-mediated inhibition of DNA synthesis and the ADP-ribosylation of Ras. ExoS was also found to cause alterations in HT-29 cell function, leading to the loss of cell adhesion and microvillus effacement. Nonadherent ExoS-treated cells remained viable but had a high proportion of modified Ras. While microvillus effacement was detected in both 388- and 388DeltaS-treated cells, effacement was more prevalent and rapid in cells exposed to strain 388. We conclude from these studies that ExoS can have multiple effects on epithelial cell function, with more severe cellular alterations associated with the enzymatic modification of Ras. The finding that ExoS had greater effects on cell growth and adherence than on cell viability suggests that ExoS may contribute to the P. aeruginosa infectious process by rendering cells nonfunctional.
Figures
FIG. 1
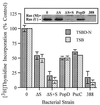
ExoS-specific effect on cell proliferation and Ras modification. DNA synthesis was examined in HT-29 cells, seeded at 105 cells/well, cultured for 48 h, and then cocultured with 107 CFU of 388, 388ΔS (ΔS), 388ΔS complemented with ExoS (ΔS+S), ΔPopD, or ΔPscC bacteria per ml (MOI of 10:1). Bacteria were either induced or noninduced for ExoS production in TSBD-N or TSB media, respectively, prior to coculture with HT-29 cells. After 4 h, bacteria were removed and replaced with medium containing [3H]thymidine and antibiotics to inhibit further bacterial growth, and DNA synthesis was assayed after 20 h. The means and standard deviations of assays performed in triplicate are indicated, and the results are expressed as percentages of non-bacterium-treated monolayer DNA synthesis. (Inset) To analyze for Ras modification, cells were cocultured in a manner identical to that described above, the culture supernatants were removed, the cells were lysed, and Ras was immunoprecipitated by using Ras monoclonal Y13-259 antibody. Proteins were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and Ras immunoblots were developed by using pan-Ras OP22 antibody and detected by using enhanced chemiluminescence. The mobility of unmodified Ras (U) and modified Ras (M) are indicated.
FIG. 2
ExoS-specific effects on HT-29 cell morphology. HT-29 cells were cocultured with strain 388, 388ΔS (ΔS), ΔS+S, ΔPopD, or ΔPscC and examined for morphological alterations by phase-contrast microscopy after 20 h. Studies were performed in conjunction with the DNA synthesis assays shown in Fig. 1.
FIG. 3
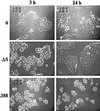
Time-lapse video analysis of effects of strains 388 and 388ΔS on HT-29 cell morphology. HT-29 cells were seeded at 5 × 105 cells/ml in 25-cm2 tissue culture flasks and grown overnight to ∼30% confluency. Cells were then cocultured with 108 CFU of 388 or 388ΔS bacteria per ml (MOI of 20:1) or no bacteria (0) for 3 h. Bacteria were removed and replaced with medium containing antibiotics. A single field of each culture was videotaped for 24 h by using a Zeiss ICM-405 inverted-phase microscope equipped with a warm stage heater-recirculator device and maintained at 37°C in a 5% CO2–95% air atmosphere. Illustrative images obtained in a single field were captured and recorded at a temporal ratio of 20:1. Magnification, ×40.
FIG. 4
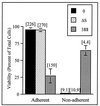
Effect of ExoS production on HT-29 cell re-adherence. HT-29 cells were cocultured with 108 CFU of bacteria per ml for 3 h (MOI of 30:1); bacteria were then removed, and cells were detached by trypsin-EDTA treatment and reseeded in culture wells. At 20 h, adherent and nonadherent cells were harvested separately, cell viability and numbers were assessed by trypan blue staining, and DNA synthesis was assayed as described in Fig. 1. Viability is expressed as the percentage of total adherent plus nonadherent cells, and the means and standard errors of three independent assays are shown. DNA synthesis, indicated in brackets above bars, is reported as the disintegrations per minute per 1,000 cells in the respective adherent or nonadherent populations. Results of one of three independent DNA assays are shown, and the mean of assays performed in triplicate is represented.
FIG. 5
Association of Ras modification with loss of adherence after exposure to ExoS-producing bacteria. HT-29 cells were cocultured with no bacteria (0) or with 108 CFU of strain 388 or strain 388ΔS (ΔS) bacteria per ml for 2, 4, or 6 h (MOI of 95:1). Bacteria were removed, and cells were reseeded in culture wells; adherent and nonadherent cells were then harvested after 20 h as described for Fig. 4. Ras was immunoprecipitated from cell lysates, and Ras modification was analyzed as described in Fig. 1 in adherent cells treated with no bacteria or with strain 388ΔS and in both adherent and nonadherent (indicated by an asterisk) cells treated with strain 388. The percentage of adherent or nonadherent viable cells in each population is indicated below the respective Ras modification patterns. The mobility of unmodified Ras (u) and modified Ras (m) is indicated.
FIG. 6
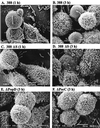
Scanning electron microscopy of HT-29 cells cocultured with ExoS-producing bacteria, non-ExoS-producing bacteria, or type III secretory mutant bacteria. Strains 388, 388ΔS, ΔPopD, and ΔPscC (MOI of ∼30:1) were cocultured for 1, 2, or 3 h with HT-29 monolayers grown on 12-mm-diameter glass coverslips. Cells were then washed, fixed in situ in 2% glutaraldehyde, postfixed in 1% osmium, dehydrated in a graded series of ethanol mixtures, and treated with hexamethyldisilane. After cells were dried, the coverslips were coated with gold and examined by using a JEOL JSEM-LV5410 scanning electron microscope. Images representative of indicated time points are shown, with each image at a ×5,000 magnification. (A and B) Cells treated with strain 388 show cell rounding and loss of cell surface microvilli after 1 and 3 h of exposure to bacteria. (C and D) Cells treated with strain 388ΔS show cell rounding after 1 and 3 h, with microvillus effacement not evident until 3 h. (E and F) Cells treated with strains ΔPopD and ΔPscC show cell rounding but no microvillus effacement after 3 h.
Similar articles
- Independent and coordinate effects of ADP-ribosyltransferase and GTPase-activating activities of exoenzyme S on HT-29 epithelial cell function.
Fraylick JE, La Rocque JR, Vincent TS, Olson JC. Fraylick JE, et al. Infect Immun. 2001 Sep;69(9):5318-28. doi: 10.1128/IAI.69.9.5318-5328.2001. Infect Immun. 2001. PMID: 11500401 Free PMC article. - ADP-ribosylation of oncogenic Ras proteins by pseudomonas aeruginosa exoenzyme S in vivo.
Vincent TS, Fraylick JE, McGuffie EM, Olson JC. Vincent TS, et al. Mol Microbiol. 1999 Jun;32(5):1054-64. doi: 10.1046/j.1365-2958.1999.01420.x. Mol Microbiol. 1999. PMID: 10361307 - Modification of Ras in eukaryotic cells by Pseudomonas aeruginosa exoenzyme S.
McGuffie EM, Frank DW, Vincent TS, Olson JC. McGuffie EM, et al. Infect Immun. 1998 Jun;66(6):2607-13. doi: 10.1128/IAI.66.6.2607-2613.1998. Infect Immun. 1998. PMID: 9596723 Free PMC article. - Pseudomonas aeruginosa exoenzyme S, a bifunctional type-III secreted cytotoxin.
Barbieri JT. Barbieri JT. Int J Med Microbiol. 2000 Oct;290(4-5):381-7. doi: 10.1016/S1438-4221(00)80047-8. Int J Med Microbiol. 2000. PMID: 11111915 Review. - The exoenzyme S regulon of Pseudomonas aeruginosa.
Frank DW. Frank DW. Mol Microbiol. 1997 Nov;26(4):621-9. doi: 10.1046/j.1365-2958.1997.6251991.x. Mol Microbiol. 1997. PMID: 9427393 Review.
Cited by
- Comparison of the exoS gene and protein expression in soil and clinical isolates of Pseudomonas aeruginosa.
Ferguson MW, Maxwell JA, Vincent TS, da Silva J, Olson JC. Ferguson MW, et al. Infect Immun. 2001 Apr;69(4):2198-210. doi: 10.1128/IAI.69.4.2198-2210.2001. Infect Immun. 2001. PMID: 11254575 Free PMC article. - Pseudomonas aeruginosa cystic fibrosis isolates induce rapid, type III secretion-dependent, but ExoU-independent, oncosis of macrophages and polymorphonuclear neutrophils.
Dacheux D, Toussaint B, Richard M, Brochier G, Croize J, Attree I. Dacheux D, et al. Infect Immun. 2000 May;68(5):2916-24. doi: 10.1128/IAI.68.5.2916-2924.2000. Infect Immun. 2000. PMID: 10768989 Free PMC article. - An indirect enzyme-linked immunosorbent assay for rapid and quantitative assessment of Type III virulence phenotypes of Pseudomonas aeruginosa isolates.
Li L, Ledizet M, Kar K, Koski RA, Kazmierczak BI. Li L, et al. Ann Clin Microbiol Antimicrob. 2005 Dec 23;4:22. doi: 10.1186/1476-0711-4-22. Ann Clin Microbiol Antimicrob. 2005. PMID: 16375761 Free PMC article. - Characterization of Pseudomonas aeruginosa exoenzyme S as a bifunctional enzyme in J774A.1 macrophages.
Rocha CL, Coburn J, Rucks EA, Olson JC. Rocha CL, et al. Infect Immun. 2003 Sep;71(9):5296-305. doi: 10.1128/IAI.71.9.5296-5305.2003. Infect Immun. 2003. PMID: 12933877 Free PMC article. - Expression of ExsA in trans confers type III secretion system-dependent cytotoxicity on noncytotoxic Pseudomonas aeruginosa cystic fibrosis isolates.
Dacheux D, Attree I, Toussaint B. Dacheux D, et al. Infect Immun. 2001 Jan;69(1):538-42. doi: 10.1128/IAI.69.1.538-542.2001. Infect Immun. 2001. PMID: 11119548 Free PMC article.
References
- Coburn J, Kane A V, Feig L, Gill D M. Pseudomonas aeruginosa exoenzyme S requires a eukaryotic protein for ADP-ribosyltransferase activity. J Biol Chem. 1991;266:6438–6446. - PubMed
- Coburn J, Wyatt R T, Iglewski B H, Gill D M. Several GTP-binding proteins, including p21c-H-ras, are preferred substrates of Pseudomonas aeruginosa exoenzyme S. J Biol Chem. 1989;264:9004–9008. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources