A single nucleotide in the SMN gene regulates splicing and is responsible for spinal muscular atrophy - PubMed (original) (raw)
A single nucleotide in the SMN gene regulates splicing and is responsible for spinal muscular atrophy
C L Lorson et al. Proc Natl Acad Sci U S A. 1999.
Abstract
SMN1 and SMN2 (survival motor neuron) encode identical proteins. A critical question is why only the homozygous loss of SMN1, and not SMN2, results in spinal muscular atrophy (SMA). Analysis of transcripts from SMN1/SMN2 hybrid genes and a new SMN1 mutation showed a direct relationship between presence of disease and exon 7 skipping. We have reported previously that the exon-skipped product SMNDelta7 is partially defective for self-association and SMN self-oligomerization correlated with clinical severity. To evaluate systematically which of the five nucleotides that differ between SMN1 and SMN2 effect alternative splicing of exon 7, a series of SMN minigenes was engineered and transfected into cultured cells, and their transcripts were characterized. Of these nucleotide differences, the exon 7 C-to-T transition at codon 280, a translationally silent variance, was necessary and sufficient to dictate exon 7 alternative splicing. Thus, the failure of SMN2 to fully compensate for SMN1 and protect from SMA is due to a nucleotide exchange (C/T) that attenuates activity of an exonic enhancer. These findings demonstrate the molecular genetic basis for the nature and pathogenesis of SMA and illustrate a novel disease mechanism. Because individuals with SMA retain the SMN2 allele, therapy targeted at preventing exon 7 skipping could modify clinical outcome.
Figures
Figure 1
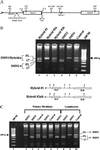
Endogenous SMN hybrid gene transcript analysis. (A) Expanded view of the 3′ region of SMN genes. Intron (lines) and exon (boxes) boundaries are indicated. Positions of the five nucleotide differences between SMN1 and SMN2 within intron 6 (In6, G/A), exon 7 (Ex7, C/T), intron 7 (In7+100, A/G; In7+214, A/G), and exon 8 (Ex8, G/A) are shown (SMN1/SMN2 sequence). The new intron 7 mutation (*In7 +6), and splice elements are shown (3′ ss, 3′ splice site; 5′ ss, 5′ splice site; PolyPy Tract, polypyrimidine tract). (B) RT-PCR amplification of SMN transcripts from Epstein–Barr virus transformed from SMA and control individuals by using oligonucleotides located in SMN exons 5 and 8. Products were digested with _Dde_I and resolved in a 2% agarose gel. The SMN2 nucleotide within exon 8 creates a _Dde_I site, resulting in faster-migrating species for SMN2 transcripts. The positions of full-length (FL) and exon 7-skipped (Δ7) transcripts are indicated. Graphic representations of the 3′ end of the hybrid genes and the origin of the five nucleotide polymorphisms are indicated (1, SMN1; 2, SMN2; In, intron; Ex, exon): Hybrid #1 (In6/SMN2, Ex7/SMN2, In7+100/SMN1, In7+214/SMN1, Ex8/SMN1); Hybrid #2a/b (In6/SMN2, Ex7/SMN2, In7+100/SMN2, In7+214/SMN2, Ex8/SMN1). (C) RT-PCR amplification of SMN transcripts from primary fibroblasts and EBV-transformed lymphocytes from SMA and control individuals (see B).
Figure 2
Analysis of plasmid-based SMN transcripts. (A) RT-PCR amplification of total RNA isolated from neuroblastoma C6 cells 48 h posttransfection with plasmid-based SMN1 (pSMN1), SMN2 (pSMN2), vector alone (pVector), or mock-transfected vector by using Lipofectamine (Life Technologies). FL SMN and SMNΔ7 transcripts are indicated and have been sequenced to ensure fidelity of splicing events. Two vector transcript species are a result of incomplete excision of the plasmid-based intron. No reverse transcriptase (-RT pSMN1) served as a control. (B) pSMN1 or pSMN2 (100 ng, 500 ng, 1 μg, 5 μg, and 10 μg) was transiently transfected into C6 cells. RT-PCR was performed (see A), and FL SMN and SMNΔ7 transcripts are indicated. (C) Transient expression of pSMN1 or pSMN2 in C33A (cervical carcinoma), C6 (neuroblastoma), T98G (glioblastoma), and Cath.a and CAD (murine neuroblastoma) cell lines and RT-PCR analysis of plasmid SMN transcripts (see A). (D) Synthetic SMN1 hybrid constructs. Two SMN2 nucleotides were introduced into a pSMN1 construct (In, intron; Ex, exon): pSMN1ΔIn6/Ex7; -ΔEx7/In7+100; -ΔEx7/In7+214. SMN2-derived nucleotides follow “Δ.” (E) Single SMN2 nucleotides introduced into pSMN1: pSMN1ΔEx7; -ΔIn6; -ΔIn7+100; -ΔIn7+214. SMN2-derived nucleotides follow “Δ.” (F) Single SMN1 nucleotides introduced into pSMN2: pSMN2ΔIn6; -ΔEx7; -ΔIn7+100. SMN2-derived nucleotides follow “Δ.” A 100-bp ladder is indicated.
Figure 3
SMNΔ7 production from a novel SMN1 spice-site mutation. RT-PCR analysis (see Fig. 1_B_) of endogenous SMN transcripts from SMA and control individuals’ EBV-transformed lymphocytes. The father is carrier-ΔSMN1, and the mother is carrierΔc.922+6 T/G. Each has an additional, single SMN1 copy. The patient carries an SMN1 deletion and an SMN1 copy with the splice-site mutation. The SMN1-derived, exon-skipped transcript is shown in lanes 5 and 6 (*Δ7).
Similar articles
- 5-(N-ethyl-N-isopropyl)-amiloride enhances SMN2 exon 7 inclusion and protein expression in spinal muscular atrophy cells.
Yuo CY, Lin HH, Chang YS, Yang WK, Chang JG. Yuo CY, et al. Ann Neurol. 2008 Jan;63(1):26-34. doi: 10.1002/ana.21241. Ann Neurol. 2008. PMID: 17924536 - Determinants of exon 7 splicing in the spinal muscular atrophy genes, SMN1 and SMN2.
Cartegni L, Hastings ML, Calarco JA, de Stanchina E, Krainer AR. Cartegni L, et al. Am J Hum Genet. 2006 Jan;78(1):63-77. doi: 10.1086/498853. Epub 2005 Nov 16. Am J Hum Genet. 2006. PMID: 16385450 Free PMC article. - SRp30c-dependent stimulation of survival motor neuron (SMN) exon 7 inclusion is facilitated by a direct interaction with hTra2 beta 1.
Young PJ, DiDonato CJ, Hu D, Kothary R, Androphy EJ, Lorson CL. Young PJ, et al. Hum Mol Genet. 2002 Mar 1;11(5):577-87. doi: 10.1093/hmg/11.5.577. Hum Mol Genet. 2002. PMID: 11875052 - Spinal muscular atrophy: from gene to therapy.
Wirth B, Brichta L, Hahnen E. Wirth B, et al. Semin Pediatr Neurol. 2006 Jun;13(2):121-31. doi: 10.1016/j.spen.2006.06.008. Semin Pediatr Neurol. 2006. PMID: 17027862 Review. - An update of the mutation spectrum of the survival motor neuron gene (SMN1) in autosomal recessive spinal muscular atrophy (SMA).
Wirth B. Wirth B. Hum Mutat. 2000;15(3):228-37. doi: 10.1002/(SICI)1098-1004(200003)15:3<228::AID-HUMU3>3.0.CO;2-9. Hum Mutat. 2000. PMID: 10679938 Review.
Cited by
- Safety, Tolerability, and Effect of Nusinersen Treatment in Ambulatory Adults With 5q-SMA.
Elsheikh B, Severyn S, Zhao S, Kline D, Linsenmayer M, Kelly K, Tellez M, Bartlett A, Heintzman S, Reynolds J, Sterling G, Weaver T, Rajneesh K, Kolb SJ, Arnold WD. Elsheikh B, et al. Front Neurol. 2021 May 20;12:650535. doi: 10.3389/fneur.2021.650535. eCollection 2021. Front Neurol. 2021. PMID: 34093395 Free PMC article. - Behavioral and electrophysiological outcomes of tissue-specific Smn knockdown in Drosophila melanogaster.
Timmerman C, Sanyal S. Timmerman C, et al. Brain Res. 2012 Dec 13;1489:66-80. doi: 10.1016/j.brainres.2012.10.035. Epub 2012 Oct 26. Brain Res. 2012. PMID: 23103409 Free PMC article. - Plastin 3 Expression Does Not Modify Spinal Muscular Atrophy Severity in the ∆7 SMA Mouse.
McGovern VL, Massoni-Laporte A, Wang X, Le TT, Le HT, Beattie CE, Rich MM, Burghes AH. McGovern VL, et al. PLoS One. 2015 Jul 2;10(7):e0132364. doi: 10.1371/journal.pone.0132364. eCollection 2015. PLoS One. 2015. PMID: 26134627 Free PMC article. - Restoration of SMN in Schwann cells reverses myelination defects and improves neuromuscular function in spinal muscular atrophy.
Hunter G, Powis RA, Jones RA, Groen EJ, Shorrock HK, Lane FM, Zheng Y, Sherman DL, Brophy PJ, Gillingwater TH. Hunter G, et al. Hum Mol Genet. 2016 Jul 1;25(13):2853-2861. doi: 10.1093/hmg/ddw141. Epub 2016 May 11. Hum Mol Genet. 2016. PMID: 27170316 Free PMC article. - Histone deacetylase inhibition suppresses myogenin-dependent atrogene activation in spinal muscular atrophy mice.
Bricceno KV, Sampognaro PJ, Van Meerbeke JP, Sumner CJ, Fischbeck KH, Burnett BG. Bricceno KV, et al. Hum Mol Genet. 2012 Oct 15;21(20):4448-59. doi: 10.1093/hmg/dds286. Epub 2012 Jul 13. Hum Mol Genet. 2012. PMID: 22798624 Free PMC article.
References
- Pearn J. Lancet. 1980;1:919–922. - PubMed
- Lefebvre S, Burglin L, Reboullet S, Clermont O, Burlet P, Viollet L, Benichou B, Cruaud C, Millasseau P, Zeviani M, et al. Cell. 1995;80:155–165. - PubMed
- Rodrigues N R, Owen N, Talbot K, Ignatius J, Dubowitz V, Davies K E. Hum Mol Genet. 1995;4:631–634. - PubMed
- Hahnen E, Forkert R, Marke C, Rudnik-Schoneborn S, Schonling J, Zerres K, Wirth B. Hum Mol Genet. 1995;4:1927–1933. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials