Characterization of MALS/Velis-1, -2, and -3: a family of mammalian LIN-7 homologs enriched at brain synapses in association with the postsynaptic density-95/NMDA receptor postsynaptic complex - PubMed (original) (raw)
Characterization of MALS/Velis-1, -2, and -3: a family of mammalian LIN-7 homologs enriched at brain synapses in association with the postsynaptic density-95/NMDA receptor postsynaptic complex
K Jo et al. J Neurosci. 1999.
Abstract
Protein assembly at the postsynaptic density (PSD) of neuronal synapses is mediated in part by protein interactions with PSD-95/discs large/zona occludens-1 (PDZ) motifs. Here, we identify MALS-1, -2, -3, a family of small synaptic proteins containing little more than a single PDZ domain. MALS-1, -2, and -3 are mammalian homologs LIN-7, a Caenorhabditis elegans protein essential for vulval development. In contrast to functions for LIN-7 in epithelial cells, MALS-1 and -2 are selectively expressed in specific neuronal populations in brain and are enriched in PSD fractions. In cultured hippocampal neurons, MALS proteins are clustered together with PSD-95 and NMDA type glutamate receptors, consistent with a postsynaptic localization for MALS proteins. Immunoprecipitation and affinity chromatography studies readily identify association of MALS with PSD-95 and an NMDA receptor subunit. The PDZ domain of MALS selectively binds to peptides terminating in E-T/S-R/X-V/I/L, which corresponds to the C terminus of NMDA type 2 receptors and numerous other ion channels at the PSD. This work suggests a role for MALS proteins in regulating recruitment of neurotransmitter receptors to the PSD.
Figures
Fig. 1.
Sequence alignment of MALS-1, -2, and -3 with LIN-7. Sequences are shown in single letter amino acid code and are aligned for maximal homology with the C. elegans LIN-7 protein. Residues present in a majority of the sequences are shaded_gray_. Arrowheads designate the PDZ domains. (GenBank accession numbers for cDNAs encoding MALS-1, -2, and -3 are AA405216, W75523, and AA462021, respectively.)
Fig. 2.
Tissue distribution of MALS-1, -2, and -3 mRNA. Northern blots of poly(A+) RNA from various rat tissues (2 μg/lane) hybridized with [32P]-labeled cDNA probes to MALS-1, -2, and -3.
Fig. 3.
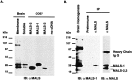
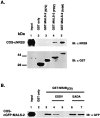
Identification of MALS proteins in brain.A, Western blotting of rat brain membrane and soluble fractions (15 μg/lane) with an affinity-purified MALS antibody demonstrates that MALS proteins are present in brain. The 25 kDa MALS band is primarily membrane-associated, and the 30 kDa MALS band is equally distributed in membrane and soluble fractions. MALS-1, -2, and -3 protein expressed in COS7 cells (3 μg/lane) comigrated with 30 (MALS-1) and 25 (MALS-2.3) kDa bands. B, Crude brain homogenate and brain extracts immunoprecipitated with preimmune serum or affinity-purified MALS antibody were immunoblotted with antiserum to MALS-1.
Fig. 4.
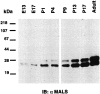
MALS proteins are selectively enriched in brain.A, Western blotting of rat tissue homogenates (15 μg/lane) probed with an affinity-purified MALS antibody demonstrates that MALS are enriched in brain. B, An immunoblot of extracts from rat brain regions (15 μg/lane) probed with the affinity-purified MALS antibody shows ubiquitous expression of MALS in rat brain. C, A sagittal section of rat brain was processed for MALS immunohistochemistry. This dark-field image suggests a neuronal localization for MALS and shows that highest levels of MALS expression occur in cerebellum (CB), cerebral cortex (CX), hippocampus (HP), striatum (ST), thalamus (T), and inferior colliculi (IC). SC, Superior colliculi; H, hypothalamus.
Fig. 5.
MALS protein expression progressively increases during brain development. Homogenates from rat brain at various developmental stages (15 μg/lane) were immunoblotted with the affinity-purified MALS antibody. Embryonic (E) and postnatal (P) days are indicated.
Fig. 6.
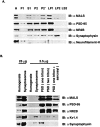
MALS proteins are enriched in PSD fractions. A, Subcellular fractions of rat cerebral cortex were prepared by differential centrifugation. Aliquots from this fractionation were probed by Western blotting with MALS antibody. Antibodies against PSD-95 and NR2B, synaptophysin and neurofilament-H, were used as controls for postsynaptic proteins, a synaptic vesicle protein and a nonsynaptic protein, respectively. Fractions (5 μg/lane, except 25 μg/lane for homogenates) are marked:H, Homogenates; P1, a crude nuclear fraction; S1, a supernatant; P2, a crude membrane fraction; P2′, a crude synaptosomal fraction;LP1, a heavy membrane fraction; LP2, a synaptic vesicle fraction; LS2, a cytosolic synaptosomal fraction. B, MALS proteins are enriched in the PSD fraction. Rat cerebral cortex homogenate and synaptosomes and isolated PSDs extracted once with 0.5% Triton X-100 (one triton), twice with 0.5% Triton X-100 (two triton), or once with 0.5% Triton X-100 followed by 3% Sarcosyl (one triton + sarcosyl) were analyzed by immunoblotting with the affinity-purified MALS antibody. Antibodies against PSD-95, NR2B and Kv1.4, and synaptophysin were used as controls for postsynaptic and presynaptic membrane proteins, respectively.
Fig. 7.
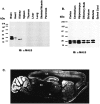
MALS is coclustered with PSD-95 and NR1 in cultured hippocampal neurons. A, Hippocampal neurons were double-labeled with NR1 (left) and MALS (right) antibodies. Arrowheads indicate apparent synaptic clusters enriched with both proteins.B, Colocalization is also apparent in cultures double-labeled for PSD-95 (left) and MALS (right) antibodies. All immunolabeling for MALS was eliminated by preabsorption of the MALS antibody with its immunogen (data not shown).
Fig. 8.
Association of MALS with PSD-95/NR2B.A, Coimmunoprecipitation of PSD-95 and NR2B with MALS. Solubilized brain membranes were immunoprecipitated with preimmune serum (lane 2) or with the affinity-purified MALS antibody (lane 3). Coimmunoprecipitation of PSD-95 and NR2B is demonstrated by immunoblotting of these immunoprecipitates. As a control, CaM kinase II α subunit was not present in the immunoprecipitates. When the extracts was presorbed with 50 μg of purified GST-MALS-2 (lane 4) or boiled with 0.5% SDS followed by addition of 5 vol of 1% Triton X-100 (lane 5), PSD-95 and NR2B were not coimmunoprecipitated. Lane 1, Input was 10% of total protein used for the immunoprecipitation. B, Coimmunoprecipitation of and NR2B and MALS with PSD-95. Solubilized brain membranes were immunoprecipitated with preimmune serum (lane 2) or with the PSD-95 antibody (lane 3). Coimmunoprecipitation of MALS and NR2B is demonstrated by immunoblotting of these immunoprecipitates. As a control, CaM kinase II α subunit was not present in the immunoprecipitates. Input, 10% (lane 1). C, Affinity chromatography demonstrates that GST:MALS-2 selectively retains PSD-95 and NR2B from solubilized cerebrocortical membranes (lane 3). Preincubating the extracts with free MALS-2 protein prevented binding of PSD-95 or NR2B to the column (lane 4). As a control, CaM kinase II α subunit was not retained by the column.D, COS7 cells were transfected with expression vectors encoding PSD-95, NR2B, or both. Cell homogenates were pulled-down with GST alone (lane 2) or with GST:MALS-2 (lane 3), and the retained proteins were analyzed by immunoblotting.
Fig. 9.
MALS binds to NR2B through a typical PDZ domain/C-terminal tail interaction. A, Solubilized extracts from COS7 cells transfected with NR2B (lane 1) were pulled-down with GST alone (lane 2), GST:MALS-2(N70) (lane 3), GST:MALS-2(PDZ) (lane 3) or GST:MALS-2(total) (lane 4). Retained proteins were analyzed by immunoblotting. B, Cell homogenates from COS7 cells transfected with GFP:MALS-2 (lane 1) were pulled-down with GST alone (lane 2) or with GST:NR2B(C9) (lanes 3–7). A peptide corresponding to the C-terminal nine amino acids of NR2B (ESDV) was added to the binding reaction at 5 (lane 4) or 50 (lane 5) μ
m
. A similar peptide in which the last four amino acids were replaced with EADA was added into the binding reaction at 5 (lane 6) or 50 (lane 7) μ
m
. Retained GFP:MALS-2 was detected by immunoblotting with a GFP antibody.
Fig. 10.
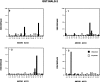
The PDZ domain of MALS-2 binds peptides terminating in E-T/S-R/X-V/I/L. Normalized amino acid abundance of the final four residues from 21 independent MALS-2 binding peptides (black bars) is compared with codon frequency in the original library (white bars).
Similar articles
- Shank, a novel family of postsynaptic density proteins that binds to the NMDA receptor/PSD-95/GKAP complex and cortactin.
Naisbitt S, Kim E, Tu JC, Xiao B, Sala C, Valtschanoff J, Weinberg RJ, Worley PF, Sheng M. Naisbitt S, et al. Neuron. 1999 Jul;23(3):569-82. doi: 10.1016/s0896-6273(00)80809-0. Neuron. 1999. PMID: 10433268 - Domain interaction between NMDA receptor subunits and the postsynaptic density protein PSD-95.
Kornau HC, Schenker LT, Kennedy MB, Seeburg PH. Kornau HC, et al. Science. 1995 Sep 22;269(5231):1737-40. doi: 10.1126/science.7569905. Science. 1995. PMID: 7569905 - Differential modulation of NR1-NR2A and NR1-NR2B subtypes of NMDA receptor by PDZ domain-containing proteins.
Iwamoto T, Yamada Y, Hori K, Watanabe Y, Sobue K, Inui M. Iwamoto T, et al. J Neurochem. 2004 Apr;89(1):100-8. doi: 10.1046/j.1471-4159.2003.02293.x. J Neurochem. 2004. PMID: 15030393 - Interactions with PDZ proteins diversify voltage-gated calcium channel signaling.
Wang S, Cortes CJ. Wang S, et al. J Neurosci Res. 2021 Jan;99(1):332-348. doi: 10.1002/jnr.24650. Epub 2020 Jun 1. J Neurosci Res. 2021. PMID: 32476168 Review. - Therapeutic use of PDZ protein-protein interaction antagonism.
Wang NX, Lee HJ, Zheng JJ. Wang NX, et al. Drug News Perspect. 2008 Apr;21(3):137-41. Drug News Perspect. 2008. PMID: 18560611 Free PMC article. Review.
Cited by
- Recent Progress on Genetically Modified Animal Models for Membrane Skeletal Proteins: The 4.1 and MPP Families.
Terada N, Saitoh Y, Saito M, Yamada T, Kamijo A, Yoshizawa T, Sakamoto T. Terada N, et al. Genes (Basel). 2023 Oct 15;14(10):1942. doi: 10.3390/genes14101942. Genes (Basel). 2023. PMID: 37895291 Free PMC article. Review. - New transmembrane AMPA receptor regulatory protein isoform, gamma-7, differentially regulates AMPA receptors.
Kato AS, Zhou W, Milstein AD, Knierman MD, Siuda ER, Dotzlaf JE, Yu H, Hale JE, Nisenbaum ES, Nicoll RA, Bredt DS. Kato AS, et al. J Neurosci. 2007 May 2;27(18):4969-77. doi: 10.1523/JNEUROSCI.5561-06.2007. J Neurosci. 2007. PMID: 17475805 Free PMC article. - PDZ protein interactions regulating glutamate receptor function and plasticity.
Tomita S, Nicoll RA, Bredt DS. Tomita S, et al. J Cell Biol. 2001 May 28;153(5):F19-24. doi: 10.1083/jcb.153.5.f19. J Cell Biol. 2001. PMID: 11381098 Free PMC article. Review. No abstract available. - SUMOylation of the MAGUK protein CASK regulates dendritic spinogenesis.
Chao HW, Hong CJ, Huang TN, Lin YL, Hsueh YP. Chao HW, et al. J Cell Biol. 2008 Jul 14;182(1):141-55. doi: 10.1083/jcb.200712094. Epub 2008 Jul 7. J Cell Biol. 2008. PMID: 18606847 Free PMC article. - Crystallization and preliminary X-ray data collection of the L27(PATJ)-(L27N,L27C)(Pals1)-L27(MALS) tripartite complex.
Zhang J, Yang X, Shen Y, Long J. Zhang J, et al. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2011 Nov 1;67(Pt 11):1443-7. doi: 10.1107/S174430911103689X. Epub 2011 Oct 27. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2011. PMID: 22102253 Free PMC article.
References
- Bredt DS. Sorting out genes that regulate epithelial and neuronal polarity. Cell. 1998;94:691–694. - PubMed
- Brenman JE, Chao DS, Xia H, Aldape K, Bredt DS. Nitric oxide synthase complexed with dystrophin and absent from skeletal muscle sarcolemma in Duchenne muscular dystrophy. Cell. 1995;82:743–752. - PubMed
- Brenman JE, Chao DS, Gee SH, McGee AW, Craven SE, Santillano DR, Huang F, Xia H, Peters MF, Froehner SC, Bredt DS. Interaction of nitric oxide synthase with the postsynaptic density protein PSD-95 and α-1 syntrophin mediated by PDZ motifs. Cell. 1996;84:757–767. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases