Analysis of fungal diversity in the wheat rhizosphere by sequencing of cloned PCR-amplified genes encoding 18S rRNA and temperature gradient gel electrophoresis - PubMed (original) (raw)
Analysis of fungal diversity in the wheat rhizosphere by sequencing of cloned PCR-amplified genes encoding 18S rRNA and temperature gradient gel electrophoresis
E Smit et al. Appl Environ Microbiol. 1999 Jun.
Abstract
Like bacteria, fungi play an important role in the soil ecosystem. As only a small fraction of the fungi present in soil can be cultured, conventional microbiological techniques yield only limited information on the composition and dynamics of fungal communities in soil. DNA-based methods do not depend on the culturability of microorganisms, and therefore they offer an attractive alternative for the study of complex fungal community structures. For this purpose, we designed various PCR primers that allow the specific amplification of fungal 18S-ribosomal-DNA (rDNA) sequences, even in the presence of nonfungal 18S rDNA. DNA was extracted from the wheat rhizosphere, and 18S rDNA gene banks were constructed in Escherichia coli by cloning PCR products generated with primer pairs EF4-EF3 (1. 4 kb) and EF4-fung5 (0.5 kb). Fragments of 0.5 kb from the cloned inserts were sequenced and compared to known rDNA sequences. Sequences from all major fungal taxa were amplified by using both primer pairs. As predicted by computer analysis, primer pair EF4-EF3 appeared slightly biased to amplify Basidiomycota and Zygomycota, whereas EF4-fung5 amplified mainly Ascomycota. The 61 clones that were sequenced matched the sequences of 24 different species in the Ribosomal Database Project (RDP) database. Similarity values ranged from 0.676 to 1. Temperature gradient gel electrophoresis (TGGE) analysis of the fungal community in the wheat rhizosphere of a microcosm experiment was carried out after amplification of total DNA with both primer pairs. This resulted in reproducible, distinctive fingerprints, confirming the difference in amplification specificity. Clear banding patterns were obtained with soil and rhizosphere samples by using both primer sets in combination. By comparing the electrophoretic mobility of community fingerprint bands to that of the bands obtained with separate clones, some could be tentatively identified. While 18S-rDNA sequences do not always provide the taxonomic resolution to identify fungal species and strains, they do provide information on the diversity and dynamics of groups of related species in environmental samples with sufficient resolution to produce discrete bands which can be separated by TGGE. This combination of 18S-rDNA PCR amplification and TGGE community analysis should allow study of the diversity, composition, and dynamics of the fungal community in bulk soil and in the rhizosphere.
Figures
FIG. 1
Primers for amplification of fungal 18S-rDNA sequences. Primer pair EF4-EF3 (fragment PCR1a) is specific for fungal 18S rDNA, and primer pair EF4-fung5 (fragment PCR1b) amplifies a smaller, 550-bp fragment specific for fungi. Primer pair EF4-NS3 (PCR2) can be used directly on clones or in a nested approach on PCR1 products for community analysis by TGGE. The primers which were developed in this work are given in alignment with 18S-rDNA sequences of species from all major fungal taxa.
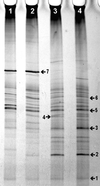
FIG. 2
TGGE of amplified 18S-rDNA fragments representing the fungal community in bulk and rhizophere soil samples of the microcosm experiment. Banding patterns were obtained by mixing the duplicate samples before PCR and adding PCR products from both primer pairs (EF4-EF3 and EF4-fung5) the same lane. Lane 1, day 5 bulk soil; lane 2, day 5 rhizosphere soil; lane 3, day 10 bulk soil; and lane 4, day 10 rhizosphere soil. Numbered bands are explained in the text.
FIG. 3
A dendrogram based on the presence or absence of bands (Fig. 2) was constructed to represent the percentages of genetic identity between the profiles of the microcosm samples of bulk and rhizosphere (rhizo) soils.
Similar articles
- Potential bias of fungal 18S rDNA and internal transcribed spacer polymerase chain reaction primers for estimating fungal biodiversity in soil.
Anderson IC, Campbell CD, Prosser JI. Anderson IC, et al. Environ Microbiol. 2003 Jan;5(1):36-47. doi: 10.1046/j.1462-2920.2003.00383.x. Environ Microbiol. 2003. PMID: 12542711 - Detection and characterization of fungal infections of Ammophila arenaria (marram grass) roots by denaturing gradient gel electrophoresis of specifically amplified 18s rDNA.
Kowalchuk GA, Gerards S, Woldendorp JW. Kowalchuk GA, et al. Appl Environ Microbiol. 1997 Oct;63(10):3858-65. doi: 10.1128/aem.63.10.3858-3865.1997. Appl Environ Microbiol. 1997. PMID: 9327549 Free PMC article. - Fungal community analysis by high-throughput sequencing of amplified markers--a user's guide.
Lindahl BD, Nilsson RH, Tedersoo L, Abarenkov K, Carlsen T, Kjøller R, Kõljalg U, Pennanen T, Rosendahl S, Stenlid J, Kauserud H. Lindahl BD, et al. New Phytol. 2013 Jul;199(1):288-299. doi: 10.1111/nph.12243. Epub 2013 Mar 28. New Phytol. 2013. PMID: 23534863 Free PMC article. Review. - Fungal taxonomy and sequence-based nomenclature.
Lücking R, Aime MC, Robbertse B, Miller AN, Aoki T, Ariyawansa HA, Cardinali G, Crous PW, Druzhinina IS, Geiser DM, Hawksworth DL, Hyde KD, Irinyi L, Jeewon R, Johnston PR, Kirk PM, Malosso E, May TW, Meyer W, Nilsson HR, Öpik M, Robert V, Stadler M, Thines M, Vu D, Yurkov AM, Zhang N, Schoch CL. Lücking R, et al. Nat Microbiol. 2021 May;6(5):540-548. doi: 10.1038/s41564-021-00888-x. Epub 2021 Apr 26. Nat Microbiol. 2021. PMID: 33903746 Free PMC article. Review.
Cited by
- Phylogenetic analysis of a spontaneous cocoa bean fermentation metagenome reveals new insights into its bacterial and fungal community diversity.
Illeghems K, De Vuyst L, Papalexandratou Z, Weckx S. Illeghems K, et al. PLoS One. 2012;7(5):e38040. doi: 10.1371/journal.pone.0038040. Epub 2012 May 29. PLoS One. 2012. PMID: 22666442 Free PMC article. - Evidence that chytrids dominate fungal communities in high-elevation soils.
Freeman KR, Martin AP, Karki D, Lynch RC, Mitter MS, Meyer AF, Longcore JE, Simmons DR, Schmidt SK. Freeman KR, et al. Proc Natl Acad Sci U S A. 2009 Oct 27;106(43):18315-20. doi: 10.1073/pnas.0907303106. Epub 2009 Oct 13. Proc Natl Acad Sci U S A. 2009. PMID: 19826082 Free PMC article. - Biodiversity and Abundance of Cultured Microfungi from the Permanently Ice-Covered Lake Fryxell, Antarctica.
Connell L, Segee B, Redman R, Rodriguez RJ, Staudigel H. Connell L, et al. Life (Basel). 2018 Sep 6;8(3):37. doi: 10.3390/life8030037. Life (Basel). 2018. PMID: 30200614 Free PMC article. - Current insights into fungal species diversity and perspective on naming the environmental DNA sequences of fungi.
Wu B, Hussain M, Zhang W, Stadler M, Liu X, Xiang M. Wu B, et al. Mycology. 2019 May 7;10(3):127-140. doi: 10.1080/21501203.2019.1614106. eCollection 2019. Mycology. 2019. PMID: 31448147 Free PMC article. Review. - Hidden mycota of pine needles: Molecular signatures from PCR-DGGE and Ribosomal DNA phylogenetic characterization of novel phylotypes.
Jeewon R, Yeung QSY, Wannasinghe DN, Rampadarath S, Puchooa D, Wang HK, Hyde KD. Jeewon R, et al. Sci Rep. 2018 Dec 21;8(1):18053. doi: 10.1038/s41598-018-36573-z. Sci Rep. 2018. PMID: 30575771 Free PMC article.
References
- Alabouvette C. Biological control of Fusarium wilt pathogens in suppressive soils. In: Hornby D, editor. Biological control of soil-borne pathogens. Wallingford, United Kingdom: CAB International; 1990. pp. 27–34.
- Bock M, Maiwald M, Kappe R, Näher H. Polymerase chain reaction-based detection of dermatophyte DNA with a fungus-specific primer system. Mycoses. 1994;37:79–84. - PubMed
- Christensen M. Species diversity and dominance in fungal communities. In: Wicklow D T, Carroll G C, editors. The fungal community. New York, N.Y: Marcel Dekker, Inc.; 1981. pp. 201–232.
- De Ley F A A M, Lynch J M. Functional diversity of the rhizosphere. In: Ogoshi A, Kobayashi K, Homma Y, Kadoma F, Kondo N, Akino S, editors. Plant growth-promoting rhizobacteria. Proceedings of the Fourth International Workshop on plant-growth promoting rhizobacteria. Paris, France: OECD; 1997. pp. 38–43.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials