Genetic analysis of a chromosomal region containing vanA and vanB, genes required for conversion of either ferulate or vanillate to protocatechuate in Acinetobacter - PubMed (original) (raw)
Genetic analysis of a chromosomal region containing vanA and vanB, genes required for conversion of either ferulate or vanillate to protocatechuate in Acinetobacter
A Segura et al. J Bacteriol. 1999 Jun.
Abstract
VanA and VanB form an oxygenative demethylase that converts vanillate to protocatechuate in microorganisms. Ferulate, an abundant phytochemical, had been shown to be metabolized through a vanillate intermediate in several Pseudomonas isolates, and biochemical evidence had indicated that vanillate also is an intermediate in ferulate catabolism by Acinetobacter. Genetic evidence supporting this conclusion was obtained by characterization of mutant Acinetobacter strains blocked in catabolism of both ferulate and vanillate. Cloned Acinetobacter vanA and vanB were shown to be members of a chromosomal segment remote from a supraoperonic cluster containing other genes required for completion of the catabolism of ferulate and its structural analogs, caffeate and coumarate, through protocatechuate. The nucleotide sequence of DNA containing vanA and vanB demonstrated the presence of genes that, on the basis of nucleotide sequence similarity, appeared to be associated with transport of aromatic compounds, metabolism of such compounds, or iron scavenging. Spontaneous deletion of 100 kb of DNA containing this segment does not impede the growth of cells with simple carbon sources other than vanillate or ferulate. Additional spontaneous mutations blocking vanA and vanB expression were shown to be mediated by IS1236, including insertion of the newly discovered composite transposon Tn5613. On the whole, vanA and vanB appear to be located within a nonessential genetic region that exhibits considerable genetic malleability in Acinetobacter. The overall organization of genes neighboring Acinetobacter vanA and vanB, including a putative transcriptional regulatory gene that is convergently transcribed and overlaps vanB, is conserved in Pseudomonas aeruginosa but has undergone radical rearrangement in other Pseudomonas species.
Figures
FIG. 1
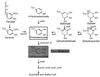
Vanillate and many other plant products are metabolized through protocatechuate. The pob, qui, and pca genes are clustered in the Acinetobacter chromosome. The open box indicates protocatechuate, and the shaded box indicates carboxymuconate, which is produced by the action of protocatechuate 3,4-dioxygenase on protocatechuate. Arrows indicate metabolic reactions and are accompanied by the designations of genes encoding the enzymes that catalyze the reactions. Metabolic accumulation of carboxymuconate in strain ADP230, defective in pcaB, prevents the growth of cells in the presence of substrates that can be metabolized to carboxymuconate.
FIG. 2
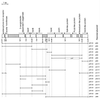
Plasmids used in this investigation. The DNA in each depicted insert is represented as a horizontal line extending between the restriction sites that mark the limits of the insert. The vectors pRK415 (39), pUC19 (66), and pBSK (Stratagene) have been described previously. All of the inserts were derived from the 14-kb _Eco_RI fragment in pZR135. The interruption in the horizontal line in pZR189 indicates the deletion produced by digestion with _Cla_I before recovery by gap repair of vanB chromosomal DNA in this plasmid. At the top of the figure are listed the functions tentatively assigned to open reading frames (indicated as open rectangles) on the basis of amino acid sequences similar to those of known proteins; the arrows indicate the directions of transcription. It must be emphasized that the some of the tentative functions assigned to proteins were shown to be incorrect as part of this investigation: a knockout mutation blocking the expression of the SalA-like protein did not prevent growth on salicylate, and the VanA-like protein did not complement mutations blocking the expression of vanA. Not depicted here are four additional _Bam_HI sites (GGATCC) that were detected by sequencing the vanA-vanK intergenic region.
FIG. 3
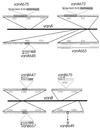
Characterized mutations in vanA and vanB. Linear bars represent vanA and vanB; expanded portions of the sequence depict the locations of mutations. Shaded rectangles indicate direct repetitions of chromosomal sequence flanking sites of insertion for mutations caused by insertion sequences or transposons. Horizontal arrows show the direction of transcription of the open reading frames within IS_1236_. Underlining marks the 7-bp direct chromosomal sequence repetition that appears to have directed the 13-bp vanA653 deletion. Characterization of DNA upstream from vanB674 was made possible by cloning of an _Eco_RI fragment containing DNA extending into the Eco_RI site of Tn_5613. Properties of DNA downstream from this site in vanB675 remains unknown.
FIG. 4
Possible contributions of DNA strand slippage to mutations in vanA. The 11-bp deletion in vanA653 removed a 7-bp direct sequence repetition, suggesting that mispairing between slipped strands may have misaligned DNA so that the deletion took place during replication. The single-stranded loop caused by mispairing may have been a target for mini-Tn_10_ insertion, resulting in vanA675.
FIG. 5
Comparative organization of the vanAB chromosomal region. From top to bottom in the figure, the percent amino acid identities to the corresponding proteins in Acinetobacter sp. strain ADP1, followed in parentheses by the numbers of aligned residues, are 69% (340), 69% (346), and 72% (340) for VanA; 46% (314), 47% (316), and 48% (316) for VanB; 61% (163), 57% (26), and 43% (166) for the regulatory protein in the GntR family (VanR); and 50% (415) for VanK. The region labelled abp could encode a protein with up to 33% identity over 148 aligned residues with a protein in various bacteria thought to be the periplasmic ATP-binding component of an ATP-binding cassette-type transport system (although this region includes a stop codon the potential open reading frame). Similarly, the size of the regulatory gene shown above assumes a frameshift in all three Pseudomonas sequences, due to mutation or sequencing error (and not included in the calculation of amino acid identity for the protein in the bottom two Pseudomonas sequences). The P. aeruginosa genes are present on one contig from the 15 September 1998 release of data from the Pseudomonas Genome Project. Arrows indicate the directions of transcription.
Similar articles
- Molecular characterization of genes of Pseudomonas sp. strain HR199 involved in bioconversion of vanillin to protocatechuate.
Priefert H, Rabenhorst J, Steinbüchel A. Priefert H, et al. J Bacteriol. 1997 Apr;179(8):2595-607. doi: 10.1128/jb.179.8.2595-2607.1997. J Bacteriol. 1997. PMID: 9098058 Free PMC article. - Bioconversion of ferulic acid into vanillic acid by means of a vanillate-negative mutant of Pseudomonas fluorescens strain BF13.
Civolani C, Barghini P, Roncetti AR, Ruzzi M, Schiesser A. Civolani C, et al. Appl Environ Microbiol. 2000 Jun;66(6):2311-7. doi: 10.1128/AEM.66.6.2311-2317.2000. Appl Environ Microbiol. 2000. PMID: 10831404 Free PMC article. - Cloning and sequencing of Pseudomonas genes encoding vanillate demethylase.
Brunel F, Davison J. Brunel F, et al. J Bacteriol. 1988 Oct;170(10):4924-30. doi: 10.1128/jb.170.10.4924-4930.1988. J Bacteriol. 1988. PMID: 3170489 Free PMC article. - The physiological contribution of Acinetobacter PcaK, a transport system that acts upon protocatechuate, can be masked by the overlapping specificity of VanK.
D'Argenio DA, Segura A, Coco WM, Bünz PV, Ornston LN. D'Argenio DA, et al. J Bacteriol. 1999 Jun;181(11):3505-15. doi: 10.1128/JB.181.11.3505-3515.1999. J Bacteriol. 1999. PMID: 10348864 Free PMC article. - Substrate range and genetic analysis of Acinetobacter vanillate demethylase.
Morawski B, Segura A, Ornston LN. Morawski B, et al. J Bacteriol. 2000 Mar;182(5):1383-9. doi: 10.1128/JB.182.5.1383-1389.2000. J Bacteriol. 2000. PMID: 10671462 Free PMC article.
Cited by
- Toxicity caused by hydroxycinnamoyl-coenzyme A thioester accumulation in mutants of Acinetobacter sp. strain ADP1.
Parke D, Ornston LN. Parke D, et al. Appl Environ Microbiol. 2004 May;70(5):2974-83. doi: 10.1128/AEM.70.5.2974-2983.2004. Appl Environ Microbiol. 2004. PMID: 15128559 Free PMC article. - A promiscuous cytochrome P450 aromatic O-demethylase for lignin bioconversion.
Mallinson SJB, Machovina MM, Silveira RL, Garcia-Borràs M, Gallup N, Johnson CW, Allen MD, Skaf MS, Crowley MF, Neidle EL, Houk KN, Beckham GT, DuBois JL, McGeehan JE. Mallinson SJB, et al. Nat Commun. 2018 Jun 27;9(1):2487. doi: 10.1038/s41467-018-04878-2. Nat Commun. 2018. PMID: 29950589 Free PMC article. - The effects of PstR, a PadR family transcriptional regulatory factor, in Plesiomonas shigelloides are revealed by transcriptomics.
Yan J, Zhang Z, Shi H, Xue X, Li A, Liu F, Ding P, Guo X, Cao B. Yan J, et al. BMC Microbiol. 2024 Nov 15;24(1):479. doi: 10.1186/s12866-024-03639-0. BMC Microbiol. 2024. PMID: 39548383 Free PMC article. - Substitution, insertion, deletion, suppression, and altered substrate specificity in functional protocatechuate 3,4-dioxygenases.
D'Argenio DA, Vetting MW, Ohlendorf DH, Ornston LN. D'Argenio DA, et al. J Bacteriol. 1999 Oct;181(20):6478-87. doi: 10.1128/JB.181.20.6478-6487.1999. J Bacteriol. 1999. PMID: 10515940 Free PMC article. - Enabling microbial syringol conversion through structure-guided protein engineering.
Machovina MM, Mallinson SJB, Knott BC, Meyers AW, Garcia-Borràs M, Bu L, Gado JE, Oliver A, Schmidt GP, Hinchen DJ, Crowley MF, Johnson CW, Neidle EL, Payne CM, Houk KN, Beckham GT, McGeehan JE, DuBois JL. Machovina MM, et al. Proc Natl Acad Sci U S A. 2019 Jul 9;116(28):13970-13976. doi: 10.1073/pnas.1820001116. Epub 2019 Jun 24. Proc Natl Acad Sci U S A. 2019. PMID: 31235604 Free PMC article.
References
- Bouvet P J, Jeanjean S. Differentiation of Acinetobacter calcoaceticus sensu stricto from related Acinetobacter species by electrophoretic polymorphism of malate dehydrogenase, glutamate dehydrogenase and catalase. Res Microbiol. 1995;146:773–785. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources