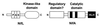PAS domains: internal sensors of oxygen, redox potential, and light - PubMed (original) (raw)
Review
PAS domains: internal sensors of oxygen, redox potential, and light
B L Taylor et al. Microbiol Mol Biol Rev. 1999 Jun.
Abstract
PAS domains are newly recognized signaling domains that are widely distributed in proteins from members of the Archaea and Bacteria and from fungi, plants, insects, and vertebrates. They function as input modules in proteins that sense oxygen, redox potential, light, and some other stimuli. Specificity in sensing arises, in part, from different cofactors that may be associated with the PAS fold. Transduction of redox signals may be a common mechanistic theme in many different PAS domains. PAS proteins are always located intracellularly but may monitor the external as well as the internal environment. One way in which prokaryotic PAS proteins sense the environment is by detecting changes in the electron transport system. This serves as an early warning system for any reduction in cellular energy levels. Human PAS proteins include hypoxia-inducible factors and voltage-sensitive ion channels; other PAS proteins are integral components of circadian clocks. Although PAS domains were only recently identified, the signaling functions with which they are associated have long been recognized as fundamental properties of living cells.
Figures
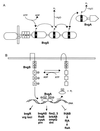
FIG. 1
Comparison of former (A) and present (B) definitions of PAS domains illustrated with the Drosophila SIM protein. (A) One PAS domain containing two PAS repeats as first described. (B) Two individual PAS domains have been identified in the SIM protein. Q-rich, glutamine-rich activation domain.
FIG. 2
Multiple alignment of PAS domains. The alignment was constructed as described in “Search strategy” (see the text) and in reference with modifications from reference . The secondary structures of PYP, FixL, and HERG were adapted from references , , and and are numbered by using the convention of Gong et al. (82). Subdomains of the crystallographic structure of PYP (172) are shown above the secondary structure. The highly variable N-terminal cap segment is not included in the alignment. Identical amino acids that are conserved in at least 50% of sequences are in reverse contrast; similar residues conserved in at least 75% of PAS domains are shaded. Consensus sequences are shown below the alignment (threshold = 75%); c, charged (DEHKR), U, bulky hydrophobic (FILMVWY); h, hydrophobic (ACFGILMTVWY); o, hydroxy (S, T); p, polar (CDEHKNQRST); t, turn-like (ACDEGHNQRST); s, small (ACDGNPSTV). An updated version of the alignment is maintained at
www.llu.edu/medicine/micro/PAS.
FIG. 2
Multiple alignment of PAS domains. The alignment was constructed as described in “Search strategy” (see the text) and in reference with modifications from reference . The secondary structures of PYP, FixL, and HERG were adapted from references , , and and are numbered by using the convention of Gong et al. (82). Subdomains of the crystallographic structure of PYP (172) are shown above the secondary structure. The highly variable N-terminal cap segment is not included in the alignment. Identical amino acids that are conserved in at least 50% of sequences are in reverse contrast; similar residues conserved in at least 75% of PAS domains are shaded. Consensus sequences are shown below the alignment (threshold = 75%); c, charged (DEHKR), U, bulky hydrophobic (FILMVWY); h, hydrophobic (ACFGILMTVWY); o, hydroxy (S, T); p, polar (CDEHKNQRST); t, turn-like (ACDEGHNQRST); s, small (ACDGNPSTV). An updated version of the alignment is maintained at
www.llu.edu/medicine/micro/PAS.
FIG. 2
Multiple alignment of PAS domains. The alignment was constructed as described in “Search strategy” (see the text) and in reference with modifications from reference . The secondary structures of PYP, FixL, and HERG were adapted from references , , and and are numbered by using the convention of Gong et al. (82). Subdomains of the crystallographic structure of PYP (172) are shown above the secondary structure. The highly variable N-terminal cap segment is not included in the alignment. Identical amino acids that are conserved in at least 50% of sequences are in reverse contrast; similar residues conserved in at least 75% of PAS domains are shaded. Consensus sequences are shown below the alignment (threshold = 75%); c, charged (DEHKR), U, bulky hydrophobic (FILMVWY); h, hydrophobic (ACFGILMTVWY); o, hydroxy (S, T); p, polar (CDEHKNQRST); t, turn-like (ACDEGHNQRST); s, small (ACDGNPSTV). An updated version of the alignment is maintained at
www.llu.edu/medicine/micro/PAS.
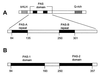
FIG. 3
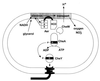
PAS domain modules in histidine kinase phosphorelay systems. (A) FixL-FixJ from Sinorhizobium meliloti. (B) Aerotaxis pathway from E. coli. (C) Pathway for initiation of sporulation in Bacillus subtilis.
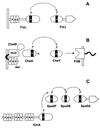
FIG. 4
PAS domain modules in photoreceptor signaling pathways and clock proteins. (A) PYP. (B) PhyA-CRY1 phosphorelay in Arabidopsis for phytochrome regulation of cryptochrome. (C) Clock protein from mouse. (D) WC-1 clock-associated protein from Neurospora crassa. Abbreviations: chr, chromophore; GATA, GATA-like zinc finger domain.
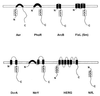
FIG. 5
Topology of selected PAS domain-containing proteins in the cytoplasmic membrane: Aer (Swiss-Prot accession no. P50466), PhoR (Swiss-Prot P08400), ArcB (Swiss-Prot P22761), FixL (PIR g628552), DcrA (Swiss-Prot P35841), NtrY (Swiss-Prot Q04850), HERG (GenBank U04270), and NifL (Swiss-Prot P30663). The putative transmembrane regions were predicted using the dense alignment surface (DAS) method (43) and comparing predictions to proteins with known topology.
FIG. 6
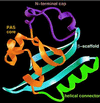
Proposed PAS three-dimensional fold illustrated on the PYP structure. The N-terminal cap (purple) includes residues 1 to 28; the PAS core (gold) includes residues 29 to 69, the helical connector (green) includes residues 70 to 87; and the β-scaffold (blue) includes residues 88 to 125. Courtesy of J. L. Pellequer and E. D. Getzoff. Reprinted from reference with permission of the publisher.
FIG. 7
Site of attachment of prosthetic groups in selected PAS domains. The sites of attachment (asterisks) are shown for 4-hydroxycinnamyl chromophore in PYP (Swiss-Prot accession no. P16133), heme b in FixL (PIR g628552), and putative [2Fe-2S] centers in NuoE (Swiss-Prot P33601). The horizontal line indicates residues that constitute the PAS core. The NuoE protein is NADH dehydrogenase I subunit E, which does not have a complete PAS domain.
FIG. 8
Domain structure of the Sll0779 protein from Synechocystis sp. strain PCC6803. The closest homolog and the closest homolog with a known function are shown for each PAS domain. Searches were performed individually with each PAS domain as a query, using the Gapped BLAST program (7). TodS, toluene sensor kinase from Pseudomonas putida (GenBank accession no. U72354); YegE, hypothetical sensor kinase from E. coli (Swiss-Prot P38097); StyS, styrene sensor kinase from Pseudomonas sp. (PID e1169869); FixL, oxygen sensor kinase from Bradyrhizobium japonicum (Swiss-Prot P23222). The hatched block represents a histidine kinase transmitter domain. Scores are given in bits. Reprinted from reference with permission of the publisher.
FIG. 9
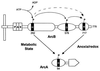
A proposed scheme for aerotaxis and redox (energy) sensing in E. coli.
FIG. 10
Schematic representation of communication modules in the ArcB-ArcA phosphorelay system. Transmembrane domains are shown in black. From the left, the modules in ArcB are PAS domain, transmitter domain, receiver domain, and HPt domain. Adapted from reference with permission of the publisher.
FIG. 11
Domain structure of NifL and NifA from Azotobacter vinelandii. Adapted from reference with permission of the publisher.
FIG. 12
Diversity of the PAS domains in the KinA protein. The closest homolog and closest homolog with known function are shown for each PAS domain. AF0456, signal transduction histidine kinase from A. fulgidis (GenBank accession no. AE001022); Arnt, aryl hydrocarbon receptor nuclear translocator from O. mykiss (GenBank U73840); DctS, dicarboxylate transport sensor kinase from R. capsulatus (Swiss-Prot P37739); HIF-1α, hypoxia-inducible factor from Mus musculus (GenBank AF045160). The methods used for this analysis are described in the legend to Fig. 8.
FIG. 13
Communication modules and phosphorelay system for control of the BvgA-BvgS regulon in Bordetella. (A) Domain structure of the BvgS sensor kinase and BvgA response regulator showing histidine kinase, phosphotransfer, and phosphotase activities associated with each domain. (B) Regulation by BvgA of the BvgA-BvgS regulon. Adapted from references and with permission of the publishers.
FIG. 14
Relatedness of PAS domains in NodV and NwsA nodulation proteins. The hatched block represents a histidine kinase transmitter domain. Black vertical bars represent transmembrane regions. The methods used for this analysis are described in the legend to Fig. 8.
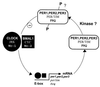
FIG. 15
A generalized circadian clock model showing the proposed role in negative feedback by proteins that contain a PAS domain. For each circadian clock component, proteins specific to mammals (top), Drosophila (middle), and Neurospora (bottom) are indicated. All proteins presented in the model, with the exception of FRQ, TIM, and the proposed kinase, contain a PAS domain. A question mark indicates hypothetical elements and events. JRK is also known as dCLOCK (4, 45).
FIG. 16
Schematic representation of the bHLH PAS-containing proteins AHR, ARNT, SIM, and HIF-1α. HIF-1α has an oxygen-dependent degradation domain that includes two PEST-like sequences, a transactivation domain (TAD), and a Fe center (104, 212). C-terminal transactivation (TAD) sites are shown in all proteins except PER. Redrawn from reference with permission of the publisher.
FIG. 17
Structural and functional organization of AHR. TAD, transactivation domain. Redrawn from reference with permission of the publisher.
Similar articles
- Structural basis for PAS domain heterodimerization in the basic helix--loop--helix-PAS transcription factor hypoxia-inducible factor.
Erbel PJ, Card PB, Karakuzu O, Bruick RK, Gardner KH. Erbel PJ, et al. Proc Natl Acad Sci U S A. 2003 Dec 23;100(26):15504-9. doi: 10.1073/pnas.2533374100. Epub 2003 Dec 10. Proc Natl Acad Sci U S A. 2003. PMID: 14668441 Free PMC article. - Molecular characterization of the murine Hif-1 alpha locus.
Luo G, Gu YZ, Jain S, Chan WK, Carr KM, Hogenesch JB, Bradfield CA. Luo G, et al. Gene Expr. 1997;6(5):287-99. Gene Expr. 1997. PMID: 9368100 Free PMC article. - Structure and function of Per-ARNT-Sim domains and their possible role in the life-cycle biology of Trypanosoma cruzi.
Rojas-Pirela M, Rigden DJ, Michels PA, Cáceres AJ, Concepción JL, Quiñones W. Rojas-Pirela M, et al. Mol Biochem Parasitol. 2018 Jan;219:52-66. doi: 10.1016/j.molbiopara.2017.11.002. Epub 2017 Nov 11. Mol Biochem Parasitol. 2018. PMID: 29133150 Review. - The mammalian basic helix-loop-helix/PAS family of transcriptional regulators.
Kewley RJ, Whitelaw ML, Chapman-Smith A. Kewley RJ, et al. Int J Biochem Cell Biol. 2004 Feb;36(2):189-204. doi: 10.1016/s1357-2725(03)00211-5. Int J Biochem Cell Biol. 2004. PMID: 14643885 Review.
Cited by
- Exploring the role of HIF-1α on pathogenesis in Alzheimer's disease and potential therapeutic approaches.
Porel P, Bala K, Aran KR. Porel P, et al. Inflammopharmacology. 2024 Oct 27. doi: 10.1007/s10787-024-01585-x. Online ahead of print. Inflammopharmacology. 2024. PMID: 39465478 Review. - Regulation of coronafacoyl phytotoxin production by the PAS-LuxR family regulator CfaR in the common scab pathogen Streptomyces scabies.
Cheng Z, Bown L, Tahlan K, Bignell DR. Cheng Z, et al. PLoS One. 2015 Mar 31;10(3):e0122450. doi: 10.1371/journal.pone.0122450. eCollection 2015. PLoS One. 2015. PMID: 25826255 Free PMC article. - The GGDEF-EAL protein CdgB from Azospirillum baldaniorum Sp245, is a dual function enzyme with potential polar localization.
Viruega-Góngora VI, Acatitla-Jácome IS, Zamorano-Sánchez D, Reyes-Carmona SR, Xiqui-Vázquez ML, Baca BE, Ramírez-Mata A. Viruega-Góngora VI, et al. PLoS One. 2022 Nov 23;17(11):e0278036. doi: 10.1371/journal.pone.0278036. eCollection 2022. PLoS One. 2022. PMID: 36417483 Free PMC article. - Cyclic Di-adenosine Monophosphate Regulates Metabolism and Growth in the Oral Commensal Streptococcus mitis.
Rørvik GH, Liskiewicz KA, Kryuchkov F, Naemi AO, Aasheim HC, Petersen FC, Küntziger TM, Simm R. Rørvik GH, et al. Microorganisms. 2020 Aug 20;8(9):1269. doi: 10.3390/microorganisms8091269. Microorganisms. 2020. PMID: 32825526 Free PMC article. - A comprehensive genetic characterization of bacterial motility.
Girgis HS, Liu Y, Ryu WS, Tavazoie S. Girgis HS, et al. PLoS Genet. 2007 Sep;3(9):1644-60. doi: 10.1371/journal.pgen.0030154. Epub 2007 Jul 25. PLoS Genet. 2007. PMID: 17941710 Free PMC article.
References
- Ahmad M, Jarillo J A, Smirnova O, Cashmore A R. The CRY1 blue light photoreceptor of Arabidopsis interacts with phytochrome A in vitro. Mol Cell. 1998;1:939–948. - PubMed
- Akerley B J, Miller J F. Understanding signal transduction during bacterial infection. Trends Microbiol. 1996;4:141–146. - PubMed
- Alex L A, Simon M I. Protein histidine kinases and signal transduction in prokaryotes and eukaryotes. Trends Genet. 1994;10:133–138. - PubMed
- Allada R, White N E, So W V, Hall J C, Rosbash M. A mutant Drosophila homolog of mammalian Clock disrupts circadian rhythms and transcription of period and timeless. Cell. 1998;93:791–804. - PubMed
- Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases