Evolution of antigen-specific T cell receptors in vivo: preimmune and antigen-driven selection of preferred complementarity-determining region 3 (CDR3) motifs - PubMed (original) (raw)
Evolution of antigen-specific T cell receptors in vivo: preimmune and antigen-driven selection of preferred complementarity-determining region 3 (CDR3) motifs
L J McHeyzer-Williams et al. J Exp Med. 1999.
Abstract
Antigen (Ag)-driven selection of helper T cells (Th) in normal animals has been difficult to study and remains poorly understood. Using the major histocompatibility complex class II- restricted murine response to pigeon cytochrome c (PCC), we provide evidence for both preimmune and Ag-driven selection in the evolution of Ag-specific immunity in vivo. Before antigenic challenge, most Valpha11(+)Vbeta3(+) Th (70%) express a critical complementarity-determining region 3 (CDR3) residue (glutamic acid at TCR-alpha93) associated with PCC peptide contact. Over the first 5 d of the primary response, PCC-responsive Valpha11(+)Vbeta3(+) Th expressing eight preferred CDR3 features are rapidly selected in vivo. Clonal dominance is further propagated through selective expansion of the PCC-specific cells with T cell receptor (TCR) of the "best fit." Ag-driven selection is complete before significant emergence of the germinal center reaction. These data argue that thymic selection shapes TCR-alpha V region bias in the preimmune repertoire; however, Ag itself and the nongerminal center microenvironment drive the selective expansion of clones with preferred TCR that dominate the response to Ag in vivo.
Figures
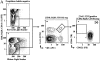
Figure 1
Five-color flow cytometric identification of PCC-specific Th. (A) Seven-parameter flow strategy outlined sequentially. Cells were stained using FITC–RR8.1 (anti-Vα11), allophycocyanin–KJ25 (anti-Vβ3), PE–Mel14 (anti-CD62L), Cy5PE–6B2 (anti-B220), Cy5PE–53-6.7 (anti-CD8), Cy5PE–M1/70 (anti-CD11b), TR–avidin/biotin–IM7 (anti-CD44), and PI as described in Materials and Methods. (i and ii) PI is excluded using the Cy5PE channel at acquisition, and cells positive for CD8, B220, and CD11b are also excluded using Cy5PE. Forward and obtuse light scatter are set to exclude many macrophages and most neutrophils but include T cell blasts. (iii) Vα11 and Vβ3 staining allows isolation of non-CD8 T cells that express both chains of the receptor shown in the insert of this panel. (iv) CD44 and CD62L levels on the T cells with TCR, using the V regions preferred for PCC specificity before injection. (B) Representative probability contours for CD44 and CD62L expression on Vα11Vβ3-expressing T cells. The day after Ag administration is displayed in the upper right of each panel, over the course of the primary response with adjuvant only (top row), primary PCC response (middle row) and memory response (bottom row). The insert box indicates the limits of CD44 upregulation and CD62L downregulation that were used to calculate frequencies of cells that respond to Ag. (C) Frequencies of Vα11Vβ3-expressing T cells that have upregulated CD44 and downregulated CD62L and total cell numbers from each animal calculated at the time animals were killed are used to estimate the change in total Ag-responsive cells in the draining LNs during the course of the primary and memory response. Varying numbers of single animals were used as indicated by the n below each timepoint on the x-axis, with means ± SEM displayed. There was no significant difference in the adjuvant-only response across different days, and it is presented together from day 3 (×2), day 5 (×2), and day 7 (×5). Significant differences (2-tail t test) are observed: in the primary response, between days 0 and 3 (P = 3.0 × 10−5), days 3 and 5 (P = 3.0 × 10−3), days 5 and 7 (P = 10−6), adjuvant-only full data set day 3 (P = 5.0 × 10−4); and in the memory response, between days 0 and 2 (P = 10−2), days 2 and 3 (P = 4.0 × 10−2).
Figure 1
Five-color flow cytometric identification of PCC-specific Th. (A) Seven-parameter flow strategy outlined sequentially. Cells were stained using FITC–RR8.1 (anti-Vα11), allophycocyanin–KJ25 (anti-Vβ3), PE–Mel14 (anti-CD62L), Cy5PE–6B2 (anti-B220), Cy5PE–53-6.7 (anti-CD8), Cy5PE–M1/70 (anti-CD11b), TR–avidin/biotin–IM7 (anti-CD44), and PI as described in Materials and Methods. (i and ii) PI is excluded using the Cy5PE channel at acquisition, and cells positive for CD8, B220, and CD11b are also excluded using Cy5PE. Forward and obtuse light scatter are set to exclude many macrophages and most neutrophils but include T cell blasts. (iii) Vα11 and Vβ3 staining allows isolation of non-CD8 T cells that express both chains of the receptor shown in the insert of this panel. (iv) CD44 and CD62L levels on the T cells with TCR, using the V regions preferred for PCC specificity before injection. (B) Representative probability contours for CD44 and CD62L expression on Vα11Vβ3-expressing T cells. The day after Ag administration is displayed in the upper right of each panel, over the course of the primary response with adjuvant only (top row), primary PCC response (middle row) and memory response (bottom row). The insert box indicates the limits of CD44 upregulation and CD62L downregulation that were used to calculate frequencies of cells that respond to Ag. (C) Frequencies of Vα11Vβ3-expressing T cells that have upregulated CD44 and downregulated CD62L and total cell numbers from each animal calculated at the time animals were killed are used to estimate the change in total Ag-responsive cells in the draining LNs during the course of the primary and memory response. Varying numbers of single animals were used as indicated by the n below each timepoint on the x-axis, with means ± SEM displayed. There was no significant difference in the adjuvant-only response across different days, and it is presented together from day 3 (×2), day 5 (×2), and day 7 (×5). Significant differences (2-tail t test) are observed: in the primary response, between days 0 and 3 (P = 3.0 × 10−5), days 3 and 5 (P = 3.0 × 10−3), days 5 and 7 (P = 10−6), adjuvant-only full data set day 3 (P = 5.0 × 10−4); and in the memory response, between days 0 and 2 (P = 10−2), days 2 and 3 (P = 4.0 × 10−2).
Figure 1
Five-color flow cytometric identification of PCC-specific Th. (A) Seven-parameter flow strategy outlined sequentially. Cells were stained using FITC–RR8.1 (anti-Vα11), allophycocyanin–KJ25 (anti-Vβ3), PE–Mel14 (anti-CD62L), Cy5PE–6B2 (anti-B220), Cy5PE–53-6.7 (anti-CD8), Cy5PE–M1/70 (anti-CD11b), TR–avidin/biotin–IM7 (anti-CD44), and PI as described in Materials and Methods. (i and ii) PI is excluded using the Cy5PE channel at acquisition, and cells positive for CD8, B220, and CD11b are also excluded using Cy5PE. Forward and obtuse light scatter are set to exclude many macrophages and most neutrophils but include T cell blasts. (iii) Vα11 and Vβ3 staining allows isolation of non-CD8 T cells that express both chains of the receptor shown in the insert of this panel. (iv) CD44 and CD62L levels on the T cells with TCR, using the V regions preferred for PCC specificity before injection. (B) Representative probability contours for CD44 and CD62L expression on Vα11Vβ3-expressing T cells. The day after Ag administration is displayed in the upper right of each panel, over the course of the primary response with adjuvant only (top row), primary PCC response (middle row) and memory response (bottom row). The insert box indicates the limits of CD44 upregulation and CD62L downregulation that were used to calculate frequencies of cells that respond to Ag. (C) Frequencies of Vα11Vβ3-expressing T cells that have upregulated CD44 and downregulated CD62L and total cell numbers from each animal calculated at the time animals were killed are used to estimate the change in total Ag-responsive cells in the draining LNs during the course of the primary and memory response. Varying numbers of single animals were used as indicated by the n below each timepoint on the x-axis, with means ± SEM displayed. There was no significant difference in the adjuvant-only response across different days, and it is presented together from day 3 (×2), day 5 (×2), and day 7 (×5). Significant differences (2-tail t test) are observed: in the primary response, between days 0 and 3 (P = 3.0 × 10−5), days 3 and 5 (P = 3.0 × 10−3), days 5 and 7 (P = 10−6), adjuvant-only full data set day 3 (P = 5.0 × 10−4); and in the memory response, between days 0 and 2 (P = 10−2), days 2 and 3 (P = 4.0 × 10−2).
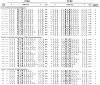
Figure 2
PCC-specific memory cells emerge with highly restricted TCR. Single PCC-specific T cells were sorted into cDNA reaction mix, subjected to two separate rounds of PCR for TCRVα11 and TCRVβ3, and then cycle-sequenced, focusing primarily on the CDR3 region of each chain from days 2 and 4 of the memory response, as indicated. A representative set of nucleotide and predicted aa sequence from single T cells where both chains of the TCR were analyzed. The TCR-α aa positions α93 and α95 and, in the TCR-β, aa positions β100 and β102, are highlighted in each sequence. The TCRJ region usage is displayed, with the CDR3 length presented between the V and J elements not considered as part of the loop (2 aa downstream from the conserved C and 2 aa upstream from the conserved GXG in the TCRJ). The N sequence in each chain is underlined, and the D region in the TCR-β chain is in bold. The preferred features column refers to the CDR3 features predominant in PCC-specific hybridoma and those which appear repeatedly in the memory response to this Ag (TCR-α: E at α93; S at α95; length of 8 aa; and TCRJα 16, 17, 22, and 34. TCR-β: N at β100; A/G at β102; length of 9 aa; and TCRJβ 1.2 and 2.5). A, alanine; E, glutamic acid; G, glycine; N, asparagine; S, serine.
Figure 3
Ag-driven selection for preferred TCR-α CDR3 features. Single-cell repertoire analysis was undertaken as described in Materials and Methods. In A–D, the CDR3 features known to be selected in the PCC response are treated separately. A, position α93; B, TCRJα usage; C, CDR3 length; and D, position α95. Each dot represents the sequence information from a single Ag-responsive cell (after day 0), with the preferred CDR3 feature presented at the top of each panel and the percentage of single cells expressing this feature displayed in the next line as “percent canonical.” Each panel organizes the information as a progression over time (from left to right) before injection (day 0, REST), after primary immunization (days 3, 5, 7, and 9 separately and 11 and 14 together), and after memory immunization (days 2, 3, and 4). The number of sequences used in the analysis are displayed as the n below the depicted days on the x-axis. The REST group includes Vα11Vβ3-expressing T cells before injection with a resting phenotype (CD44loCD62Lhi) (n = 40) and those from days 3 and 5, also with the resting phenotype (n = 22). Both groups contained similarly diverse TCR-α CDR3 regions, as did the few cells sampled from the adjuvant-only set on day 7 after primary injection (not shown). (E) A summary of the change in the percentage of single cells that expressed ≥3 of the preferred TCR-α CDR3 features over time during the response to PCC. These data represent a percentage for each animal analyzed (where there were ≥10 sequences) and the mean ± SEM for each timepoint (n below the x-axis, number of animals from which sequences were taken; n = 308 individual sequences used for the analysis). Significant differences (2-tail t test) were observed between days 0 and 3 (P = 0.01) and days 3 and 5 (P = 0.01).
Figure 4
Ag-driven selection for preferred TCR-β CDR3 features. Single-cell repertoire analysis was undertaken as described in Materials and Methods. In sections A–D, the CDR3 features known to be selected in the PCC response are treated separately. (A) Position β100, (B) TCRJβ usage, (C) CDR3 length, and (D) position β102. Each dot represents the sequence information from a single, Ag-responsive cell (after day 0), and the information is organized as described in the Fig. 3 legend. The REST group includes Vα11Vβ3-expressing T cells before injection, with a resting phenotype (CD44loCD62Lhi) (n = 48) and those from days 3 and 5, also with the resting phenotype (n = 33). Both groups contained similarly diverse TCR-β CDR3 regions, as did the few cells sampled from the adjuvant-only set on day 7 after primary injection (not shown). (E) A summary of the change in the percentage of single cells that expressed ≥3 of the preferred TCR-β CDR3 motifs over time during the response to PCC. These data represent a percentage for each animal analyzed (where there were ≥10 sequences) and the mean ± SEM for each timepoint (n below the x-axis represents the number of animals, whereas the n = 485 within the panel is the number of individual sequences used for the analysis). Significant differences (2-tail t test) were observed between days 0 and 3 (P = 0.04) and days 3 and 5 (P = 0.01).
Figure 4
Ag-driven selection for preferred TCR-β CDR3 features. Single-cell repertoire analysis was undertaken as described in Materials and Methods. In sections A–D, the CDR3 features known to be selected in the PCC response are treated separately. (A) Position β100, (B) TCRJβ usage, (C) CDR3 length, and (D) position β102. Each dot represents the sequence information from a single, Ag-responsive cell (after day 0), and the information is organized as described in the Fig. 3 legend. The REST group includes Vα11Vβ3-expressing T cells before injection, with a resting phenotype (CD44loCD62Lhi) (n = 48) and those from days 3 and 5, also with the resting phenotype (n = 33). Both groups contained similarly diverse TCR-β CDR3 regions, as did the few cells sampled from the adjuvant-only set on day 7 after primary injection (not shown). (E) A summary of the change in the percentage of single cells that expressed ≥3 of the preferred TCR-β CDR3 motifs over time during the response to PCC. These data represent a percentage for each animal analyzed (where there were ≥10 sequences) and the mean ± SEM for each timepoint (n below the x-axis represents the number of animals, whereas the n = 485 within the panel is the number of individual sequences used for the analysis). Significant differences (2-tail t test) were observed between days 0 and 3 (P = 0.04) and days 3 and 5 (P = 0.01).
Figure 4
Ag-driven selection for preferred TCR-β CDR3 features. Single-cell repertoire analysis was undertaken as described in Materials and Methods. In sections A–D, the CDR3 features known to be selected in the PCC response are treated separately. (A) Position β100, (B) TCRJβ usage, (C) CDR3 length, and (D) position β102. Each dot represents the sequence information from a single, Ag-responsive cell (after day 0), and the information is organized as described in the Fig. 3 legend. The REST group includes Vα11Vβ3-expressing T cells before injection, with a resting phenotype (CD44loCD62Lhi) (n = 48) and those from days 3 and 5, also with the resting phenotype (n = 33). Both groups contained similarly diverse TCR-β CDR3 regions, as did the few cells sampled from the adjuvant-only set on day 7 after primary injection (not shown). (E) A summary of the change in the percentage of single cells that expressed ≥3 of the preferred TCR-β CDR3 motifs over time during the response to PCC. These data represent a percentage for each animal analyzed (where there were ≥10 sequences) and the mean ± SEM for each timepoint (n below the x-axis represents the number of animals, whereas the n = 485 within the panel is the number of individual sequences used for the analysis). Significant differences (2-tail t test) were observed between days 0 and 3 (P = 0.04) and days 3 and 5 (P = 0.01).
Figure 4
Ag-driven selection for preferred TCR-β CDR3 features. Single-cell repertoire analysis was undertaken as described in Materials and Methods. In sections A–D, the CDR3 features known to be selected in the PCC response are treated separately. (A) Position β100, (B) TCRJβ usage, (C) CDR3 length, and (D) position β102. Each dot represents the sequence information from a single, Ag-responsive cell (after day 0), and the information is organized as described in the Fig. 3 legend. The REST group includes Vα11Vβ3-expressing T cells before injection, with a resting phenotype (CD44loCD62Lhi) (n = 48) and those from days 3 and 5, also with the resting phenotype (n = 33). Both groups contained similarly diverse TCR-β CDR3 regions, as did the few cells sampled from the adjuvant-only set on day 7 after primary injection (not shown). (E) A summary of the change in the percentage of single cells that expressed ≥3 of the preferred TCR-β CDR3 motifs over time during the response to PCC. These data represent a percentage for each animal analyzed (where there were ≥10 sequences) and the mean ± SEM for each timepoint (n below the x-axis represents the number of animals, whereas the n = 485 within the panel is the number of individual sequences used for the analysis). Significant differences (2-tail t test) were observed between days 0 and 3 (P = 0.04) and days 3 and 5 (P = 0.01).
Figure 4
Ag-driven selection for preferred TCR-β CDR3 features. Single-cell repertoire analysis was undertaken as described in Materials and Methods. In sections A–D, the CDR3 features known to be selected in the PCC response are treated separately. (A) Position β100, (B) TCRJβ usage, (C) CDR3 length, and (D) position β102. Each dot represents the sequence information from a single, Ag-responsive cell (after day 0), and the information is organized as described in the Fig. 3 legend. The REST group includes Vα11Vβ3-expressing T cells before injection, with a resting phenotype (CD44loCD62Lhi) (n = 48) and those from days 3 and 5, also with the resting phenotype (n = 33). Both groups contained similarly diverse TCR-β CDR3 regions, as did the few cells sampled from the adjuvant-only set on day 7 after primary injection (not shown). (E) A summary of the change in the percentage of single cells that expressed ≥3 of the preferred TCR-β CDR3 motifs over time during the response to PCC. These data represent a percentage for each animal analyzed (where there were ≥10 sequences) and the mean ± SEM for each timepoint (n below the x-axis represents the number of animals, whereas the n = 485 within the panel is the number of individual sequences used for the analysis). Significant differences (2-tail t test) were observed between days 0 and 3 (P = 0.04) and days 3 and 5 (P = 0.01).
Figure 5
Ag-driven selection for preferred CDR3 features in both chains of TCR. Single-cell repertoire analysis was undertaken as described in Materials and Methods. Each dot represents the sequence information of a single cell from which both TCR-α and TCR-β were sequenced. The y-axis represents the number of preferred CDR3 features seen in each cell. Cells with ≥6 preferred features were considered as having restricted TCR, and the percentage of these cells is displayed as part of the figure (there was very little difference in the distribution of cells after day 5 of the primary response and, therefore, they were grouped to display similar n for each group). Sequence information from the memory response cells across days 2, 3, 4, and 6 was pooled, as these cells also displayed very little difference in the distribution of preferred features. The number of sequences used in the analysis are displayed as the n on the x-axis.
Figure 6
CDR3 diversity on day 3 of the primary response. Representative CDR3 sequence data from single, PCC-responsive T cells on day 3 of the primary response is displayed in three groups. The sequence is organized as described in Fig. 2, with TCR-α and TCR-β from each cell presented across the figure. The TCR-α aa positions α93 and α95 and the TCR-β aa positions β100 and β102 are highlighted in each sequence. Group 1 represents sequences considered to have restricted TCR (≥6 preferred features). Group 2 represents sequences that appear unrestricted (≤5 preferred CDR3 features) but where the TCR-α have at least three motifs in common with a known PCC-specific hybridoma (11, 14, 17, 32, 50) or tetramer-binding cells (tet) (McHeyzer-Williams, L.J., J.F. Panus, J.A. Mikszta, J.D. Altman, M.M. Davis, and M.G. McHeyzer-Williams, manuscript in preparation). Group 3 represents unrestricted sequences (≤5 preferred CDR3 features) that bear no resemblance to previously sequenced hybridomas in either chain of the TCR.
Figure 7
GC and non-GC distribution of PCC-specific Th. (A) Cryosections were prepared from the draining LNs of B10.BR mice 7 d after initial priming and stained with FITC–RR8.1 (anti-Vα11; green), allophycocyanin-KJ25 (anti-Vβ3; red), and TR-11.26 (anti-IgD; cyan). Single-color versions of the same image were collected separately (and serially in the primary detector) using LSCM analysis (40× objective lens), and then processed, colorized, and reassembled using Adobe Photoshop. IgD is used to locate the T zones (right) and B zones and GC (left panel). The cells expressing Vα11 and Vβ3 double-stain (yellow) and are more concentrated in the GC than the T zones. (B) Comparisons of Vα11Vβ3 frequencies obtained by flow cytometric analysis of total LN cells (100,000 event files; mean for four separate experiments ± SEM) and frequencies obtained by LSCM analysis (full cross-sections through LN; mean for four full sections from three separate animals ± SEM). Total numbers of Vα11 and/or Vβ3 cells are considered 100%, with the table displaying frequencies for single- and double-positive cells by each mode of analysis. (C) Summary for the distribution of Vα11Vβ3-expressing T cells over time and in three distinct locations across the draining LN. Also displayed is the total number of cells counted across three animals for each timepoint with the displayed phenotype, across three to four full cross-sections of LN. For each timepoint, the three animals pooled display little difference in distribution. (D) These day 7 primary response cryosections were stained with FITC–Mel14 (anti-CD62L; green), allophycocyanin-KJ25 (anti-Vβ3; red), and TR-11.26 (anti-IgD; not displayed. IgD− outline of a GC is represented as a dashed circle, left). Vβ3-expressing T cells in the GC (the majority of which are also Vα11+ in serial sections) are negative for CD62L (red in the GC; 40× objective lens), whereas the majority of T zone Vβ3+ cells coexpress CD62L (right, yellow; 63× objective lens).
Figure 7
GC and non-GC distribution of PCC-specific Th. (A) Cryosections were prepared from the draining LNs of B10.BR mice 7 d after initial priming and stained with FITC–RR8.1 (anti-Vα11; green), allophycocyanin-KJ25 (anti-Vβ3; red), and TR-11.26 (anti-IgD; cyan). Single-color versions of the same image were collected separately (and serially in the primary detector) using LSCM analysis (40× objective lens), and then processed, colorized, and reassembled using Adobe Photoshop. IgD is used to locate the T zones (right) and B zones and GC (left panel). The cells expressing Vα11 and Vβ3 double-stain (yellow) and are more concentrated in the GC than the T zones. (B) Comparisons of Vα11Vβ3 frequencies obtained by flow cytometric analysis of total LN cells (100,000 event files; mean for four separate experiments ± SEM) and frequencies obtained by LSCM analysis (full cross-sections through LN; mean for four full sections from three separate animals ± SEM). Total numbers of Vα11 and/or Vβ3 cells are considered 100%, with the table displaying frequencies for single- and double-positive cells by each mode of analysis. (C) Summary for the distribution of Vα11Vβ3-expressing T cells over time and in three distinct locations across the draining LN. Also displayed is the total number of cells counted across three animals for each timepoint with the displayed phenotype, across three to four full cross-sections of LN. For each timepoint, the three animals pooled display little difference in distribution. (D) These day 7 primary response cryosections were stained with FITC–Mel14 (anti-CD62L; green), allophycocyanin-KJ25 (anti-Vβ3; red), and TR-11.26 (anti-IgD; not displayed. IgD− outline of a GC is represented as a dashed circle, left). Vβ3-expressing T cells in the GC (the majority of which are also Vα11+ in serial sections) are negative for CD62L (red in the GC; 40× objective lens), whereas the majority of T zone Vβ3+ cells coexpress CD62L (right, yellow; 63× objective lens).
Figure 7
GC and non-GC distribution of PCC-specific Th. (A) Cryosections were prepared from the draining LNs of B10.BR mice 7 d after initial priming and stained with FITC–RR8.1 (anti-Vα11; green), allophycocyanin-KJ25 (anti-Vβ3; red), and TR-11.26 (anti-IgD; cyan). Single-color versions of the same image were collected separately (and serially in the primary detector) using LSCM analysis (40× objective lens), and then processed, colorized, and reassembled using Adobe Photoshop. IgD is used to locate the T zones (right) and B zones and GC (left panel). The cells expressing Vα11 and Vβ3 double-stain (yellow) and are more concentrated in the GC than the T zones. (B) Comparisons of Vα11Vβ3 frequencies obtained by flow cytometric analysis of total LN cells (100,000 event files; mean for four separate experiments ± SEM) and frequencies obtained by LSCM analysis (full cross-sections through LN; mean for four full sections from three separate animals ± SEM). Total numbers of Vα11 and/or Vβ3 cells are considered 100%, with the table displaying frequencies for single- and double-positive cells by each mode of analysis. (C) Summary for the distribution of Vα11Vβ3-expressing T cells over time and in three distinct locations across the draining LN. Also displayed is the total number of cells counted across three animals for each timepoint with the displayed phenotype, across three to four full cross-sections of LN. For each timepoint, the three animals pooled display little difference in distribution. (D) These day 7 primary response cryosections were stained with FITC–Mel14 (anti-CD62L; green), allophycocyanin-KJ25 (anti-Vβ3; red), and TR-11.26 (anti-IgD; not displayed. IgD− outline of a GC is represented as a dashed circle, left). Vβ3-expressing T cells in the GC (the majority of which are also Vα11+ in serial sections) are negative for CD62L (red in the GC; 40× objective lens), whereas the majority of T zone Vβ3+ cells coexpress CD62L (right, yellow; 63× objective lens).
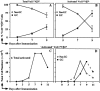
Figure 8
A shift from non-GC to GC pathway in the second week after priming. (A) Change in GC and non-GC distribution over days 5, 7, and 9 of the primary PCC response (as shown in Fig. 7 C) without adjusting for the number of Vα11Vβ3-expressing cells that do not downregulate CD62L in response to immunization. (B) The trends in Fig. 8 A have been adjusted for mean percentage of CD62LloVα11Vβ3 cells at each timepoint determined by flow cytometry (13 ± 5.1% on day 5, n = 13; 43 ± 9.4% on day 7, n = 11; and 40 ± 12% on day 9, n = 12), assuming that all GC T cells are CD62Llo (Fig. 7). (C) Change in total activated cells over the course of the primary response (means from Fig. 1 C) on a linear scale to emphasize the decline phase of the response. (D) Application of the frequencies shown in Fig. 8 B to estimate the change in total cell numbers in each pathway over the course of the primary response (GC are absent on day 0. In situ studies were not performed for days 3 and 11; however, to display the trends, we extrapolated day 5 frequencies to day 3 and day 9 frequencies to day 11).
Similar articles
- Antigen-specific T helper cell function: differential cytokine expression in primary and memory responses.
Panus JF, McHeyzer-Williams LJ, McHeyzer-Williams MG. Panus JF, et al. J Exp Med. 2000 Nov 6;192(9):1301-16. doi: 10.1084/jem.192.9.1301. J Exp Med. 2000. PMID: 11067879 Free PMC article. - Antigen-driven selection of TCR In vivo: related TCR alpha-chains pair with diverse TCR beta-chains.
Mikszta JA, McHeyzer-Williams LJ, McHeyzer-Williams MG. Mikszta JA, et al. J Immunol. 1999 Dec 1;163(11):5978-88. J Immunol. 1999. PMID: 10570285 - Two distinct mechanisms account for the immune response (Ir) gene control of the T cell response to pigeon cytochrome c.
McElligott DL, Sorger SB, Matis LA, Hedrick SM. McElligott DL, et al. J Immunol. 1988 Jun 15;140(12):4123-31. J Immunol. 1988. PMID: 2453567 - Enumeration and characterization of memory cells in the TH compartment.
McHeyzer-Williams MG, Altman JD, Davis MM. McHeyzer-Williams MG, et al. Immunol Rev. 1996 Apr;150:5-21. doi: 10.1111/j.1600-065x.1996.tb00693.x. Immunol Rev. 1996. PMID: 8782699 Review. - T-cell receptor V-region usage and antigen specificity. The cytochrome c model system.
Davis MM, McHeyzer-Williams M, Chien YH. Davis MM, et al. Ann N Y Acad Sci. 1995 Jul 7;756:1-11. doi: 10.1111/j.1749-6632.1995.tb44477.x. Ann N Y Acad Sci. 1995. PMID: 7645810 Review.
Cited by
- Machine Learning Approaches to TCR Repertoire Analysis.
Katayama Y, Yokota R, Akiyama T, Kobayashi TJ. Katayama Y, et al. Front Immunol. 2022 Jul 15;13:858057. doi: 10.3389/fimmu.2022.858057. eCollection 2022. Front Immunol. 2022. PMID: 35911778 Free PMC article. Review. - CD8+ T cell immunodominance shifts during the early stages of acute LCMV infection independently from functional avidity maturation.
Raué HP, Slifka MK. Raué HP, et al. Virology. 2009 Aug 1;390(2):197-204. doi: 10.1016/j.virol.2009.05.021. Epub 2009 Jun 18. Virology. 2009. PMID: 19539966 Free PMC article. - CD4+ T cells with convergent TCR recombination reprogram stroma and halt tumor progression in adoptive therapy.
Wolf SP, Leisegang M, Steiner M, Wallace V, Kiyotani K, Hu Y, Rosenberger L, Huang J, Schreiber K, Nakamura Y, Schietinger A, Schreiber H. Wolf SP, et al. Sci Immunol. 2024 Sep 13;9(99):eadp6529. doi: 10.1126/sciimmunol.adp6529. Epub 2024 Sep 13. Sci Immunol. 2024. PMID: 39270007 Free PMC article. - Antigen-specific T helper cell function: differential cytokine expression in primary and memory responses.
Panus JF, McHeyzer-Williams LJ, McHeyzer-Williams MG. Panus JF, et al. J Exp Med. 2000 Nov 6;192(9):1301-16. doi: 10.1084/jem.192.9.1301. J Exp Med. 2000. PMID: 11067879 Free PMC article. - Sizing up the key determinants of the CD8(+) T cell response.
Tscharke DC, Croft NP, Doherty PC, La Gruta NL. Tscharke DC, et al. Nat Rev Immunol. 2015 Nov;15(11):705-16. doi: 10.1038/nri3905. Epub 2015 Oct 9. Nat Rev Immunol. 2015. PMID: 26449178 Review.
References
- Davis MM. T cell receptor gene diversity and selection. Annu Rev Biochem. 1990;59:475–496. - PubMed
- Bevan MJ. In a radiation chimaera, host H-2 antigens determine immune responsiveness of donor cytotoxic cells. Nature. 1977;269:417–418. - PubMed
- Zinkernagel RM, Doherty PC. Restriction of in vitro T cell-mediated cytotoxicity in lymphocytic choriomeningitis within a syngeneic or semiallogeneic system. Nature. 1974;248:701–702. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous