Somatic cell mutants resistant to retrovirus replication: intracellular blocks during the early stages of infection - PubMed (original) (raw)
Somatic cell mutants resistant to retrovirus replication: intracellular blocks during the early stages of infection
G Gao et al. Mol Biol Cell. 1999 Jun.
Free PMC article
Abstract
To identify cellular functions involved in the early phase of the retroviral life cycle, somatic cell mutants were isolated after selection for resistance to infection. Rat2 fibroblasts were treated with chemical mutagens, and individual virus-resistant clones were recovered after selection for resistance to infection. Two clones were characterized in detail. Both mutant lines were resistant to infection by both ecotropic and amphotropic murine viruses, as well as by human immunodeficiency virus type 1 pseudotypes. One clone showed a strong block to reverse transcription of the retroviral RNA, including formation of the earliest DNA products. The second clone showed normal levels of viral DNA synthesis but did not allow formation of the circular DNAs normally found in the nucleus. Cell fractionation showed that the viral preintegration complex was present in a form that could not be extracted under conditions that readily extracted the complex from wild-type cells. The results suggest that the DNA was trapped in a nonproductive state and excluded from the nucleus of the infected cell. The properties of these two mutant lines suggest that host gene products play important roles both before and after reverse transcription.
Figures
Figure 1
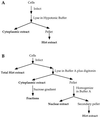
Schematic representation of the strategy for isolating mutant cells resistant to retroviral infection. TK−, thymidine kinase deficient; TK+, thymidine kinase proficient.
Figure 2
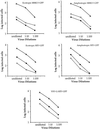
Resistance of mutant cells to GFP virus infection. RC-2 cells (open triangles), R3-2 cells (closed circles), and R4-7 cells (open circles) were infected with the indicated viruses at indicated dilution. The number of infected cells, as indicated by expression of GFP, was determined by flow cytometry.
Figure 3
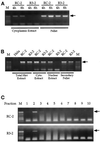
Analysis of viral DNA in infected mutant cells. RC-2 and mutant cells were infected with various dilutions of virus, and the amount of viral DNA synthesized in the infected cells was measured by PCR under conditions controlled such that the yield of PCR product was proportional to the input DNA. (A) Cells were infected with MuLV-GFP virus at various dilutions. At different time points the viral DNA in the infected cells was extracted and detected by PCR using primers that specifically amplify the GFP sequence. (B) The same preparations at the 24-h postinfection time point in A were used as templates to detect the synthesis of plus strand viral DNA in the infected cells. The migration position of the PCR product is indicated. (C) Cells were infected with either ecotropic MuLV-GFP or ecotropic MuLV-N2 virus at various dilutions, as indicated. Twenty four hours after infection the cells were collected, and minus strand strong stop DNA was detected by PCR. The migration position of the PCR product is indicated. (D) RC-2 and R4-7 cells were infected with amphotropic MuLV-GFP virus at various dilutions. Twenty-four hours after infection, viral DNA synthesis was analyzed by PCR using primers that amplify GFP sequences. (E) Cells were infected with VSV-G–pseudotyped HIV-GFP virus at various dilutions, as indicated. At 24 h after infection, cells were collected, and minus strand strong stop DNA and plus strand DNA were detected by PCR. The migration positions of the PCR products are indicated. H.I. virus, heat-inactivated virus was used as a control for plasmid DNA contamination of the virus. (F) The same preparations from A were analyzed by PCR using primers that specifically amplify the LTR–LTR junction to detect circular DNAs in the nucleus.
Figure 4
Schematic representation of the procedure for fractionating infected cells. RC-2 and R3-2 cells were infected with undiluted ecotropic MuLV-GFP virus and extracted with either hypotonic buffer (A) or buffer containing digitonin (B).
Figure 5
Analysis of viral DNA in the fractionated cell extracts by PCR using primers that amplify GFP sequences. The migration positions of the PCR product are indicated with arrows. (A) Four or 8 h after infection cells were collected and fractionated. The low-molecular-weight DNA in the pellet (Hirt extract) was resuspended in 20 μl of Tris-EDTA plus 10 μg/ml RNase A. Five of 300 μl of cytoplasmic extract or 1 of 20 μl of the pellet DNA were PCR amplified to detect viral DNA. (B) Eight hours after infection the cells were collected by trypsinization. One-third of the cells were used to extract total DNA by the Hirt procedure. The rest of the cells were fractionated as described in Figure 4B. Five of 300 μl of each extract or 1 of 60 μl of the total DNA were used to assay viral DNA by PCR. (C) The cytoplasmic extracts from B were fractionated by centrifugation on a 20–70% sucrose gradient. The gradient was equally divided into 10 fractions of 500 μl each. Five microliters of solution from each fraction were used to detect viral DNA by PCR.
Similar articles
- Isolation and characterization of human cells resistant to retrovirus infection.
Lech P, Somia NV. Lech P, et al. Retrovirology. 2007 Jul 3;4:45. doi: 10.1186/1742-4690-4-45. Retrovirology. 2007. PMID: 17608937 Free PMC article. - Isolation of cell lines that show novel, murine leukemia virus-specific blocks to early steps of retroviral replication.
Bruce JW, Bradley KA, Ahlquist P, Young JA. Bruce JW, et al. J Virol. 2005 Oct;79(20):12969-78. doi: 10.1128/JVI.79.20.12969-12978.2005. J Virol. 2005. PMID: 16188999 Free PMC article. - Intracellular trafficking of retroviral genomes during the early phase of infection: viral exploitation of cellular pathways.
Goff SP. Goff SP. J Gene Med. 2001 Nov-Dec;3(6):517-28. doi: 10.1002/1521-2254(200111)3:6<517::AID-JGM234>3.0.CO;2-E. J Gene Med. 2001. PMID: 11778899 Review. - Retrovirus restriction factors.
Goff SP. Goff SP. Mol Cell. 2004 Dec 22;16(6):849-59. doi: 10.1016/j.molcel.2004.12.001. Mol Cell. 2004. PMID: 15610729 Review.
Cited by
- Time-resolved imaging of HIV-1 Env-mediated lipid and content mixing between a single virion and cell membrane.
Markosyan RM, Cohen FS, Melikyan GB. Markosyan RM, et al. Mol Biol Cell. 2005 Dec;16(12):5502-13. doi: 10.1091/mbc.e05-06-0496. Epub 2005 Sep 29. Mol Biol Cell. 2005. PMID: 16195349 Free PMC article. - Isolation of suppressor genes that restore retrovirus susceptibility to a virus-resistant cell line.
Gao G, Goff SP. Gao G, et al. Retrovirology. 2004 Sep 28;1:30. doi: 10.1186/1742-4690-1-30. Retrovirology. 2004. PMID: 15453907 Free PMC article. - Reconstitution of retroviral fusion and uncoating in a cell-free system.
Narayan S, Young JA. Narayan S, et al. Proc Natl Acad Sci U S A. 2004 May 18;101(20):7721-6. doi: 10.1073/pnas.0401312101. Epub 2004 May 5. Proc Natl Acad Sci U S A. 2004. PMID: 15128947 Free PMC article. - HIV-1 infection is blocked at an early stage in cells devoid of mitochondrial DNA.
Lu G, Matsuura SE, Barrientos A, Scott WA. Lu G, et al. PLoS One. 2013 Oct 21;8(10):e78035. doi: 10.1371/journal.pone.0078035. eCollection 2013. PLoS One. 2013. PMID: 24205077 Free PMC article. - A phenotypic recessive, post-entry block in rabbit cells that results in aberrant trafficking of HIV-1.
Cutiño-Moguel T, Fassati A. Cutiño-Moguel T, et al. Traffic. 2006 Aug;7(8):978-92. doi: 10.1111/j.1600-0854.2006.00449.x. Traffic. 2006. PMID: 16882040 Free PMC article.
References
- Alin K, Goff SP. Amino acid substitutions in the CA protein of Moloney murine leukemia virus that block early events in infection. Virology. 1996;222:339–351. - PubMed
- Best S, Le TP, Towers G, Stoye JP. Positional cloning of the mouse retrovirus restriction gene Fv1. Nature. 1996;382:826–829. - PubMed
- Bowerman B, Brown PO, Bishop JM, Varmus HE. A nucleoprotein complex mediates the integration of retroviral DNA. Genes & Dev. 1989;3:469–478. - PubMed
- Brown PO, Bowerman B, Varmus HE, Bishop JM. Correct integration of retroviral DNA in vitro. Cell. 1987;49:347–356. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources