ZEB represses transcription through interaction with the corepressor CtBP - PubMed (original) (raw)
ZEB represses transcription through interaction with the corepressor CtBP
A A Postigo et al. Proc Natl Acad Sci U S A. 1999.
Abstract
ZEB is an active transcriptional repressor that regulates lymphocyte and muscle differentiation in vertebrates. Its homologue in Drosophila (zfh-1) is also essential for differentiation of somatic and cardiac muscle. Here, we demonstrate that ZEB and zfh-1 interact with the corepressor CtBP to repress transcription. ZEB and zfh-1, both contain the sequence PLDLS in the same region of the repressor domain, and we demonstrate that this sequence binds CtBP-1 and -2. In vertebrate species, ZEB contains two additional CtBP-like binding sites (variations of the PLDLS sequence) that also bind CtBP proteins and are required for full repressor activity. The three sites have an additive effect, and mutation of all three sites is necessary to abolish both binding to CtBP and repressor activity. Finally, we demonstrate that the interaction of CtBP with ZEB at the promoter is necessary for repressor activity.
Figures
Figure 1
ZEB and zfh-1 bind to CtBP-1 and CtBP-2. (A) ZEB and zfh-1 contain CtBP binding sites. Drosophila zfh-1 and ZEB from various vertebrate species (h: human; ck: chicken; hms: hamster; ms: mouse and rat) contain a PLDLS CtBP binding sequence in their repressor domain. In addition, vertebrates contain two additional PLDLS-like sequences, which vary slightly from species to species. (B) Flag-tagged constructs for snail, the repressor domains of ZEB and zfh-1 (the region between both zinc finger domains) as well as the DNA binding domains (C-terminal zinc fingers) of ZEB (DB-ZEB) and zfh-1 (DB-zfh-1) were cotransfected in C33a cells with myc-tagged CtBP-1. After 48 hr, cells were lysed and after immunoprecipitation with Flag antibody, binding to CtBP-1 was detected by Western blot using 9E10 anti-myc mAb as described in Materials and Methods. Ten percent of the lysate was run without immunoprecipitation as input control. Blots then were stripped and incubated with anti-Flag antibody to detect levels of snail, ZEB and zfh-1. (C) As in B but using myc-tagged CtBP-2.
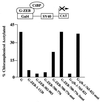
Figure 2
Binding of ZEB to CtBP mediates transcriptional repression. The region of ZEB corresponding to amino acids 1–1120, 302–903, and 700–776 were fused to the DNA binding domain of the yeast Gal4 protein and tested for its ability to repress transcription of the SV40 promoter/enhancer. Two micrograms of the Gal4 PM1 empty vector, 2 μg of wild-type G-ZEB-700–776 (amino acids 700–776), 2 μg of G-ZEB-700–776-mut (amino acids 700–776 with mutation of the CtBP site at 734), 2 μg of G-ZEB3 mut (amino acids 700–776 with mutation of CtBP sites at 705, 734, and 767), 3 μg of G-ZEB-302–903 (amino acids 302–903), 4 μg of G-ZEB-1–1120 (full-length ZEB), 2 μg of wild-type G-_zfh_-_1_–765-821 (amino acids 765–821) and 2 μg of G-_zfh-1_–765-821-mut (amino acids 765–821 with a mutation of the CtBP site at 792) were cotransfected with 0.8 μg of a reporter containing the SV40 enhancer/promoter (18). Transfection and assessment of CAT activity was performed as described in Materials and Methods.
Figure 3
(A) Mutation of all three CtBP binding sites in ZEB abolishes binding to CtBP. Flag-tagged expression vectors for snail, the DNA binding domain of ZEB, and the region of ZEB between amino acids 700 and 776 (either wild type or mutated in all three CtBP binding sites, as described in Fig. 2) were cotransfected along with myc-tagged CtBP-1. Cells were collected, and binding to CtBP-1 was detected by Western blot using 9E10 anti-myc mAb as described in Materials and Methods. The blot then was stripped and reprobed with anti-Flag antibody to check expression levels of the Flag proteins. (B) Consensus binding sequence for CtBP. Amino acids at positions −2 and +1 are constant in all proteins and species (outlined). The residue at position 0 is also highly conserved with Asp and Asn in most cases. The residues at positions −1 and +2 show great variability. The size of the residue indicates the frequency for the presence of that residue. The consensus sequence was established by accounting all proteins so far known to interact with CtBP proteins: E1A regions of adenovirus types 2, 5, and 12, CtIP, ZEB in all species where it has been cloned (see Fig. 1_A_), BKLF, zfh-1, hairy, snail, and knirps.
Figure 4
CtBP and ZEB must interact at the promoter to repress transcription. Soluble ZEB (amino acids 700–776, without the Gal4 DNA binding domain) is not able to repress transcription of the reporter containing Gal4 binding sites upstream of the SV40 promoter/enhancer. Two micrograms of either G-ZEB (amino acids 700–776 of ZEB fused to Gal4 binding domain), G-ZEB-3 mut (amino acids 700–776 of ZEB fused to Gal4 binding domain but with all three CtBP sites at positions 705, 734, and 767 mutated) or soluble ZEB were cotransfected with 0.8 μg of the SV40 promoter/enhancer reporter. However, overexpression of soluble ZEB and soluble snail inhibit repression by G-ZEB (amino acids 700–776) bound to the promoter. Increasing amounts of the soluble ZEB expression vector (2.3 and 4.7 μg) or soluble snail expression vector (6.2 μg) were cotransfected with 0.8 μg of a SV40 enhancer/promoter reporter and 2 μg of the G-ZEB (amino acids 700–776) expression vector.
Similar articles
- Differential expression and function of members of the zfh-1 family of zinc finger/homeodomain repressors.
Postigo AA, Dean DC. Postigo AA, et al. Proc Natl Acad Sci U S A. 2000 Jun 6;97(12):6391-6. doi: 10.1073/pnas.97.12.6391. Proc Natl Acad Sci U S A. 2000. PMID: 10841546 Free PMC article. - zfh-1, the Drosophila homologue of ZEB, is a transcriptional repressor that regulates somatic myogenesis.
Postigo AA, Ward E, Skeath JB, Dean DC. Postigo AA, et al. Mol Cell Biol. 1999 Oct;19(10):7255-63. doi: 10.1128/MCB.19.10.7255. Mol Cell Biol. 1999. PMID: 10490660 Free PMC article. - Interaction of ZEB and histone deacetylase with the PLDLS-binding cleft region of monomeric C-terminal binding protein 2.
Zhao LJ, Kuppuswamy M, Vijayalingam S, Chinnadurai G. Zhao LJ, et al. BMC Mol Biol. 2009 Sep 15;10:89. doi: 10.1186/1471-2199-10-89. BMC Mol Biol. 2009. PMID: 19754958 Free PMC article. - Transcriptional regulation by C-terminal binding proteins.
Chinnadurai G. Chinnadurai G. Int J Biochem Cell Biol. 2007;39(9):1593-607. doi: 10.1016/j.biocel.2007.01.025. Epub 2007 Feb 4. Int J Biochem Cell Biol. 2007. PMID: 17336131 Review. - CtBP, an unconventional transcriptional corepressor in development and oncogenesis.
Chinnadurai G. Chinnadurai G. Mol Cell. 2002 Feb;9(2):213-24. doi: 10.1016/s1097-2765(02)00443-4. Mol Cell. 2002. PMID: 11864595 Review.
Cited by
- Interpretation of allele-specific chromatin accessibility using cell state-aware deep learning.
Atak ZK, Taskiran II, Demeulemeester J, Flerin C, Mauduit D, Minnoye L, Hulselmans G, Christiaens V, Ghanem GE, Wouters J, Aerts S. Atak ZK, et al. Genome Res. 2021 Jun;31(6):1082-1096. doi: 10.1101/gr.260851.120. Epub 2021 Apr 8. Genome Res. 2021. PMID: 33832990 Free PMC article. - The miR-200 family and its targets regulate type II cell differentiation in human fetal lung.
Benlhabib H, Guo W, Pierce BM, Mendelson CR. Benlhabib H, et al. J Biol Chem. 2015 Sep 11;290(37):22409-22. doi: 10.1074/jbc.M114.636068. Epub 2015 Jul 22. J Biol Chem. 2015. PMID: 26203191 Free PMC article. - The Post-Translational Regulation of Epithelial-Mesenchymal Transition-Inducing Transcription Factors in Cancer Metastasis.
Kang E, Seo J, Yoon H, Cho S. Kang E, et al. Int J Mol Sci. 2021 Mar 30;22(7):3591. doi: 10.3390/ijms22073591. Int J Mol Sci. 2021. PMID: 33808323 Free PMC article. Review. - LncRNA SNHG14/miR-5590-3p/ZEB1 positive feedback loop promoted diffuse large B cell lymphoma progression and immune evasion through regulating PD-1/PD-L1 checkpoint.
Zhao L, Liu Y, Zhang J, Liu Y, Qi Q. Zhao L, et al. Cell Death Dis. 2019 Sep 30;10(10):731. doi: 10.1038/s41419-019-1886-5. Cell Death Dis. 2019. PMID: 31570691 Free PMC article. - Establishment of distinct MyoD, E2A, and twist DNA binding specificities by different basic region-DNA conformations.
Kophengnavong T, Michnowicz JE, Blackwell TK. Kophengnavong T, et al. Mol Cell Biol. 2000 Jan;20(1):261-72. doi: 10.1128/MCB.20.1.261-272.2000. Mol Cell Biol. 2000. PMID: 10594029 Free PMC article.
References
- Gray S, Levine M. Curr Opin Cell Biol. 1998;8:358–364. - PubMed
- Eckner R. Biol Chem. 1996;377:685–688. - PubMed
- Alland L, Muhle R, Hou H, Potes J, Chin L, Schreiber-Agus N, DePinho R A. Nature (London) 1997;387:49–55. - PubMed
- Luo R X, Postigo A A, Dean D D. Cell. 1998;92:463–473. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases