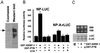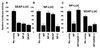Regulation of eukaryotic protein synthesis: selective influenza viral mRNA translation is mediated by the cellular RNA-binding protein GRSF-1 - PubMed (original) (raw)
Regulation of eukaryotic protein synthesis: selective influenza viral mRNA translation is mediated by the cellular RNA-binding protein GRSF-1
Y W Park et al. Proc Natl Acad Sci U S A. 1999.
Abstract
To better understand regulation of eukaryotic protein synthesis, we studied cellular and viral mRNA translation in influenza virus-infected cells. Influenza virus infection results in a dramatic shut-off of cellular protein synthesis that is concomitant with selective viral mRNA translation. Earlier work showed that these events are mediated by viral and/or cellular factors binding to the 5' untranslated region (5' UTR) of viral mRNAs. To identify trans-acting cellular proteins responsible for selective viral protein synthesis, we employed the yeast three-hybrid system. Using the 5' UTR of the influenza virus nucleocapsid protein (NP) mRNA as bait, we identified the cellular RNA-recognition motif containing RNA-binding protein G-rich sequence factor 1 (GRSF-1) as a positive-acting translational regulatory factor. The in vivo yeast assay revealed GRSF-1 specifically bound to the NP 5' UTR but not select NP 5' UTR mutants or cellular RNA 5' UTRs. These data were confirmed by gel shift assays using recombinant GRSF-1. Importantly, recombinant GRSF-1 specifically stimulated translation of a NP 5' UTR-driven template in cell-free translation systems. Furthermore, translation efficiency of NP 5' UTR-driven templates was reduced markedly in GRSF-1-depleted HeLa cell extracts, but restored in GRSF-1-reconstituted extracts. GRSF-1 also stimulated translation of an NP 5' UTR-driven template in HeLa cell extracts that were depleted of essential factors by addition of RNA oligonucleotides representing the viral 5' UTR RNA. Taken together, these data document the functional demonstration of a cellular protein binding to influenza virus RNAs and, importantly, suggest that influenza virus may recruit GRSF-1 to the 5' UTR to ensure preferential translation of viral mRNAs in infected cells.
Figures
Figure 1
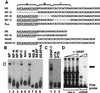
Yeast three-hybrid analysis reveals that GRSF-1 binds to influenza virus NP 5′ UTR. (A) Sequence and computer-predicted structure of the wild-type and mutant bait RNAs. The 5′ UTR of influenza viral NP mRNA or its 12-nt deletion mutant (NP-A) was placed at the 3′ end of the tandem MS2 RNAs. (B) Specific interaction of clone 149 with the wild-type bait RNA. The plasmid isolated from clone 149 was reintroduced into yeast strain L40-coat with the plasmid expressing the wild-type (NP), mutant (NP-A), or negative control (IRE) bait RNA. Yeast double transformants were assayed for β-galactosidase activity by direct measurement of enzyme activity. (C) Diagrammatic representation of a partial listing of RNA-binding proteins that share similar domain organization to the RRM-containing GRSF-1 protein is presented.
Figure 2
GRSF-1 interacts with specific sequences within the influenza virus mRNA 5′ UTR as detected by gel mobility-shift analysis. (A) Sequences of the in vitro transcribed RNA transcripts used as probes in gel mobility-shift assays. Underlined sequences represent the conserved 12-nt sequences found on all influenza virus type A mRNAs. The 5′ UTR of NP mRNA was divided into three regions (regions A, B, and C) as depicted across the top. The name of each transcript is indicated on the left side of its sequence. AINV is a derivative of NP, in which region A is reversed. The NS 5′ UTR is shown along with a sequence of the substitution mutant NS-B (mutated bases are shown in lowercase letters). Below are shown the sequences of the SEAP 5′ UTR along with the SEAP 5′ UTR appended to region A (SEAP+A). (B) Recombinant GRSF-1 (0.050 μg) purified from E. coli was incubated in the presence of the nonspecific competitor, heparin (0.125 mg/ml), with the various probes indicated across the top, as described under Materials and Methods. (C) The RRM-containing RNA-binding purified proteins U1A and PTB (0.050 μg) were incubated with the NP 5′ UTR probe as specificity controls (for details, see B). (D) HeLa S10 extract (200 μg, Left) or recombinant GRSF-1 (rGRSF-1) (0.050 μg, Right) was incubated with the NP 5′ UTR in the presence of increasing amounts of monoclonal anti-GRSF-1 for supershift analysis. The resulting RNA–protein complexes were resolved on a native polyacrylamide gel. The GRSF-1-RNA complex (open arrow on the left) and its antibody complex (solid arrow on the right) are indicated.
Figure 3
GRSF-1 selectively enhances translation of the influenza viral NP 5′ UTR-driven template. (A) GST-GRSF-1 fusion proteins (0.200 μg) were visualized by Coomassie staining or Western blotting. The arrow indicates the intact GST-GRSF-1 fusion protein. (B) The NP 5′ UTR-driven (NP-LUC) and mutant NP 5′ UTR-driven (NP-A-rLUC) templates (0.125 μg each) were translated in a HeLa extract in the absence or the presence of GST-GRSF-1 (0.200 μg). GST-PTB fusion protein (0.200 μg) and U1A (0.200 μg) again were utilized as specificity controls. After 45 min at 30°C, translation products were assayed by using a Dual-Luciferase Reporter Assay System (Promega). Values are the mean ± SD of three experiments per group. A scintillation counter was used to measure luciferase activity. Counts per minute (cpm) were produced by calculating the square root of measured cpm minus background cpm. We arbitrarily assigned a value of 100 to the control NP-luciferase reaction that, in this case, represented an average value of 3,560 cpm of luciferase activity per μl of HeLa extract. Other relative luciferase activity values were calculated relative to this number. (C) GST-GRSF-1 does not affect stability of template RNAs. Aliquots of translation products in B were extracted with phenol and phenol/chloroform, and RNAs were fractionated by formaldehyde-agarose gel electrophoresis. After electrophoresis, gels were stained with ethidium bromide to visualize ribosomal RNAs (18S and 28S, Upper) for internal controls. Template RNAs, which had been radiolabeled with trace amounts of 32P, then were visualized on x-ray film after gels were dried. LUC and rLUC indicate the firefly luciferase and the sea pansy luciferase RNA, respectively. M indicates mixture of an aliquot of the starting material of templates only.
Figure 4
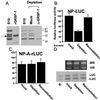
Immunodepletion of GRSF-1 compromises translation of influenza virus 5′ UTR-driven chimeric mRNA translation, whereas GRSF-1 reconstitution restores mRNA translation. (A) HeLa S10 extracts (S10) were incubated with protein A-agarose beads coated with GRSF-1-specific IgG molecules (anti-GRSF-1) or with normal mouse IgG molecules (Mock) at 4°C for 4 hr. Depleted extracts then were centrifuged briefly, and the supernatant was examined by Western blotting. The GRSF-1 isoforms in the S10 starting material (100 μg) and the migration of the recombinant GRSF-1 (50 ng) are shown on the left. The GRSF-1 proteins patterned after depletion are shown on the right. (B and C) The mock-depleted, GRSF-1-depleted, or GRSF-1-reconstituted extracts were used for the cell-free translation of wild-type and mutant NP 5′ UTR-driven mRNA translation [NP-LUC (B) and NP-A-rLUC (C)]. For the GRSF-1-reconstituted extracts, 0.200 μg of GST-GRSF-1 fusion protein was added. After 45 min at 30°C, translation products were assayed by using a Dual-Luciferase Reporter Assay System (Promega). In this experiment, 100 was equivalent to an average of 2,340 cpm of luciferase activity per μl of HeLa extract. Mock, Depletion, or Reconstitution at the bottom indicates the mock-depleted, GRSF-1-depleted, or GRSF-1-reconstituted extract, respectively. Values are the mean ± SD of three experiments per group. (D) GST-GRSF-1 does not affect stability of template RNAs. Using aliquots (12 μl) of translation products (B and C), template RNA stability was tested as described in Fig. 3_C_. LUC and rLUC indicate the firefly luciferase and the sea pansy luciferase RNA, respectively. M indicates mixture of an aliquot of the starting-material templates.
Figure 5
RNA oligonucleotide-competition experiments confirm a GRSF-1-positive translational regulatory function. (A and B) RNA oligonucleotide-competition experiments. Template cellular 5′ UTR SEAP-LUC (A) or viral 5′ UTR NP-LUC (B) was translated in an influenza virus-infected HeLa cell extract in the absence or presence of various competitors as indicated below each bar. Competitor RNAs were added at 200 × molar ratio to template. The nucleotide sequences of competitors are shown in Fig. 2_A_. An arbitrary value of 100 represented 4,215 cpm SEAP-LUC of luciferase activity per μl of HeLa cell extract in A and 4,520 cpm of NP-LUC luciferase activity per μl of HeLa cell extract in B. (C) Effects of GRSF-1 reconstitution in RNA oligonucleotide-compromised extracts. Either template NP-LUC or SEAP-LUC was translated in a virus-infected HeLa extract in the absence (No comp) or presence of competitor NP 5′ UTRs (NP) or in the presence of competitor NP 5′ UTRs plus 0.200 μg of GST-GRSF-1 (NP+GRSF-1) as indicated below each bar. After 45 min at 30°C, translation products were assayed by using a Luciferase Assay System. A value of 100 represented 4,240 cpm of NP-LUC luciferase activity per μl of HeLa cell extract in C. Other values then were determined relative to this standard. Values throughout are the mean ± SD of three experiments per group.
Similar articles
- Selective translation of eukaryotic mRNAs: functional molecular analysis of GRSF-1, a positive regulator of influenza virus protein synthesis.
Kash JC, Cunningham DM, Smit MW, Park Y, Fritz D, Wilusz J, Katze MG. Kash JC, et al. J Virol. 2002 Oct;76(20):10417-26. doi: 10.1128/jvi.76.20.10417-10426.2002. J Virol. 2002. PMID: 12239318 Free PMC article. - Functional Analysis of GRSF1 in the Nuclear Export and Translation of Influenza A Virus mRNAs.
Schmierer J, Takimoto T. Schmierer J, et al. Viruses. 2024 Jul 16;16(7):1136. doi: 10.3390/v16071136. Viruses. 2024. PMID: 39066299 Free PMC article. - So similar, yet so different: selective translation of capped and polyadenylated viral mRNAs in the influenza virus infected cell.
Yángüez E, Nieto A. Yángüez E, et al. Virus Res. 2011 Mar;156(1-2):1-12. doi: 10.1016/j.virusres.2010.12.016. Epub 2010 Dec 31. Virus Res. 2011. PMID: 21195735 Review. - The role of the 5' untranslated region of an mRNA in translation regulation during development.
van der Velden AW, Thomas AA. van der Velden AW, et al. Int J Biochem Cell Biol. 1999 Jan;31(1):87-106. doi: 10.1016/s1357-2725(98)00134-4. Int J Biochem Cell Biol. 1999. PMID: 10216946 Review.
Cited by
- Research resource: identification of novel coregulators specific for thyroid hormone receptor-β2.
Hahm JB, Privalsky ML. Hahm JB, et al. Mol Endocrinol. 2013 May;27(5):840-59. doi: 10.1210/me.2012-1117. Epub 2013 Apr 4. Mol Endocrinol. 2013. PMID: 23558175 Free PMC article. - Structural basis of G-tract recognition and encaging by hnRNP F quasi-RRMs.
Dominguez C, Fisette JF, Chabot B, Allain FH. Dominguez C, et al. Nat Struct Mol Biol. 2010 Jul;17(7):853-61. doi: 10.1038/nsmb.1814. Epub 2010 Jun 6. Nat Struct Mol Biol. 2010. PMID: 20526337 - Impact of the segment-specific region of the 3'-untranslated region of the influenza A virus PB1 segment on protein expression.
Ma J, Liu K, Xue C, Zhou J, Xu S, Ren Y, Zheng J, Cao Y. Ma J, et al. Virus Genes. 2013 Dec;47(3):429-38. doi: 10.1007/s11262-013-0969-0. Epub 2013 Aug 15. Virus Genes. 2013. PMID: 23949786 - Systems-based candidate genes for human response to influenza infection.
Zhang L, Katz JM, Gwinn M, Dowling NF, Khoury MJ. Zhang L, et al. Infect Genet Evol. 2009 Dec;9(6):1148-57. doi: 10.1016/j.meegid.2009.07.006. Epub 2009 Jul 30. Infect Genet Evol. 2009. PMID: 19647099 Free PMC article. Review. - Male guanine-rich RNA sequence binding factor 1 knockout mice (Grsf1-/-) gain less body weight during adolescence and adulthood.
Dumoulin B, Heydeck D, Jähn D, Lassé M, Sofi S, Ufer C, Kuhn H. Dumoulin B, et al. Cell Biosci. 2022 Dec 9;12(1):199. doi: 10.1186/s13578-022-00922-3. Cell Biosci. 2022. PMID: 36494688 Free PMC article.
References
- Mathews M B. In: Interactions Between Viruses and the Cellular Machinery for Protein Synthesis. Hershey J, Mathews M B, Sonenberg N, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1996. pp. 505–548.
- Ehrenfeld E. In: Initiation of Translation by Picornavirus RNAs. Hershey J, Mathews M B, Sonenberg N, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1996. pp. 549–574.
- Hinnebusch A G. In: Translational Control of GCN4: Gene-Specific Regulation by Phosphorylation of eIF2. Hershey J, Mathews M, Sonenberg N, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1996. pp. 199–244.
- Katze M G. In: Translational Control in Cells Infected with Influenza Virus and Reovirus. Hershey J W B, Mathews M B, Sonenberg N, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1996. pp. 607–630.
- Merrick W C, Hershey J W B. In: The Pathway and Mechanism of Eukaryotic Protein Synthesis. Hershey J, Mathews M, Sonenberg N, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1996. pp. 31–70.
Publication types
MeSH terms
Substances
Grants and funding
- P51 RR000166/RR/NCRR NIH HHS/United States
- RR-00166/RR/NCRR NIH HHS/United States
- GM-56434/GM/NIGMS NIH HHS/United States
- AI-22646/AI/NIAID NIH HHS/United States
- R01 AI022646/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Miscellaneous