Unexpected abundance of self-splicing introns in the genome of bacteriophage Twort: introns in multiple genes, a single gene with three introns, and exon skipping by group I ribozymes - PubMed (original) (raw)
Unexpected abundance of self-splicing introns in the genome of bacteriophage Twort: introns in multiple genes, a single gene with three introns, and exon skipping by group I ribozymes
M Landthaler et al. Proc Natl Acad Sci U S A. 1999.
Abstract
Analysis of RNA that can be labeled with GTP indicates the existence of group I introns in genes of at least three transcriptional classes in the genome of Staphylococcus aureus bacteriophage Twort. A single ORF of 142 amino acids (Orf142) is interrupted by three self-splicing group I introns, providing the first example of a phage gene with multiple intron insertions. Twort Orf142 is encoded in a message that is abundant 15-20 min after infection and is highly similar to a late gene product (Orf8) of the morphologically related Listeria phage A511. The introns in orf142 are spliced in vivo and contain all the conserved features of primary sequence and secondary structure of group I introns in subgroup IA2, which includes the introns in Escherichia coli phage T4 and the Bacillus phages beta22 and SPO1. Introns I2 and I3 in orf142 are highly similar, and their intron insertion sites are closely spaced. The presence of transcripts with a skipped exon between these introns indicates that they may fold into a single active ribozyme resulting in alternative splicing. Alternatively, the cleaved 5' exon preceding I2 may undergo trans splicing to the 3' exon that follows I3. Regardless of the detailed mechanism, these results demonstrate a new means whereby a single gene can give rise to multiple messenger RNAs.
Figures
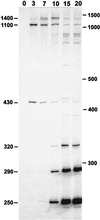
Figure 1
In vitro [α-32P]GTP labeling of S. aureus bacteriophage Twort RNA. RNA (2.5 μg) isolated from S. aureus before (0) and at times indicated after infection with Twort was deproteinized and incubated with [α-32P]GTP under self-splicing conditions. Samples were separated by electrophoresis in a 4% acrylamide/6 M urea gel. Approximate sizes of the major GTP-labeled products are indicated on the left of the autoradiogram. Positions of the 100-bp ladder size standards are indicated to the right.
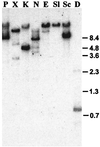
Figure 2
Southern blot hybridization of GTP-labeled Twort RNA to genomic DNA. Twort DNA was digested with _Pst_I (P), _Xba_I (X), _Kpn_I (K), _Nsi_I (N), _Eco_RI (E), _Sal_I (Sl), _Sac_I (Sc), and _Dra_I (D) and transferred. The blot was probed with 50 μg 15-min Twort RNA that was end-labeled with [α-32P] GTP under self-splicing conditions. Sizes of _Bst_EII digested λ DNA fragments are indicated to the right.
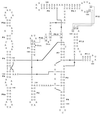
Figure 3
Genomic organization and restriction map. Relevant restriction sites are given. Predicted ORFs are shown as boxes. orfa and orfc are incomplete. Introns in gene orf142 are shown as half-sized boxes. Positions of the oligonucleotides used for PCR and RT-PCR are represented with arrows, with the arrowhead indicating the 3′ end of the primer. Location of the cloned DNA fragments is shown and plasmid names are given.
Figure 4
Nucleotide sequence of the cloned region of the Twort genome. Predicted amino acid sequences are given below. Stop codons are indicated with asterisks. Presumed ribosomal binding sites (RBS) and restriction sites are underlined. Inverted repeat downstream of orf142 is indicated by arrows. Intron splice sites are shown as downward-pointing arrows.
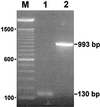
Figure 5
In vivo splicing. Electrophoretic analysis of (1) RT-PCR product obtained by using primers 4 and 5 on 15-min Twort RNA and (2) PCR product obtained by using the same primers on Twort phage DNA. Sizes of the PCR products are indicated to the right. A 100-bp ladder was used as a marker (M), with sizes given on the left.
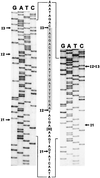
Figure 6
orf142 splice junctions. The sequencing ladders of the major 130-bp RT-PCR product (Left) and the minor 107-bp RT-PCR product (Right) around the intron insertion sites are shown. Lanes are labeled with the dideoxynucleotides. The sequence of the ligated exons is given between the ladders (with 30 nt of exon 2 omitted). The shaded sequence (exon 3) is deleted in the 107-bp RT-PCR product. Arrows indicate the intron insertion sites.
Figure 7
Sequence alignment of Twort orf142 introns and T4 nrdB intron. Conserved base-paired regions (P1-P9) are underlined. Numbers in brackets stand for nucleotides not included in the alignment. Vertical arrows point out the 5′ and 3′ splice sites. Lowercase letters indicate exon sequence and uppercase letters indicate intron sequence.
Figure 8
Secondary structure of group I intron _orf142_-I2. Exon sequence in lowercase and intron sequence in uppercase letters. Arrows indicate 5′ and 3′ splice sites (s.s.). Conserved structural elements P1 through P10 are indicated, and putative tertiary interaction P12 is shown (23).
Similar articles
- Two self-splicing group I introns in the ribonucleotide reductase large subunit gene of Staphylococcus aureus phage Twort.
Landthaler M, Begley U, Lau NC, Shub DA. Landthaler M, et al. Nucleic Acids Res. 2002 May 1;30(9):1935-43. doi: 10.1093/nar/30.9.1935. Nucleic Acids Res. 2002. PMID: 11972330 Free PMC article. - Two self-splicing group I introns interrupt two late transcribed genes of prolate-headed Lactobacillus delbrueckii phage JCL1032.
Riipinen KA, Alatossava T. Riipinen KA, et al. Arch Virol. 2004 Oct;149(10):2013-24. doi: 10.1007/s00705-004-0329-4. Arch Virol. 2004. PMID: 15669111 - Crystal structure of a phage Twort group I ribozyme-product complex.
Golden BL, Kim H, Chase E. Golden BL, et al. Nat Struct Mol Biol. 2005 Jan;12(1):82-9. doi: 10.1038/nsmb868. Epub 2004 Dec 5. Nat Struct Mol Biol. 2005. PMID: 15580277 - Genomics of staphylococcal Twort-like phages--potential therapeutics of the post-antibiotic era.
Łobocka M, Hejnowicz MS, Dąbrowski K, Gozdek A, Kosakowski J, Witkowska M, Ulatowska MI, Weber-Dąbrowska B, Kwiatek M, Parasion S, Gawor J, Kosowska H, Głowacka A. Łobocka M, et al. Adv Virus Res. 2012;83:143-216. doi: 10.1016/B978-0-12-394438-2.00005-0. Adv Virus Res. 2012. PMID: 22748811 Review. - RNA splicing in the T-even bacteriophage.
Chu FK, Maley GF, Maley F. Chu FK, et al. FASEB J. 1988 Mar 1;2(3):216-23. doi: 10.1096/fasebj.2.3.3280375. FASEB J. 1988. PMID: 3280375 Review.
Cited by
- Development of advanced control material for reverse transcription-mediated bacterial nucleic acid amplification tests.
Lewis JD, Salipante SJ. Lewis JD, et al. J Clin Microbiol. 2024 May 8;62(5):e0024324. doi: 10.1128/jcm.00243-24. Epub 2024 Apr 17. J Clin Microbiol. 2024. PMID: 38629844 Free PMC article. - Group I Intron as a Potential Target for Antifungal Compounds: Development of a _Trans_-Splicing High-Throughput Screening Strategy.
Malbert B, Labaurie V, Dorme C, Paget E. Malbert B, et al. Molecules. 2023 May 31;28(11):4460. doi: 10.3390/molecules28114460. Molecules. 2023. PMID: 37298936 Free PMC article. - Pairwise Engineering of Tandemly Aligned Self-Splicing Group I Introns for Analysis and Control of Their Alternative Splicing.
Ueda T, Nishimura KI, Nishiyama Y, Tominaga Y, Miyazaki K, Furuta H, Matsumura S, Ikawa Y. Ueda T, et al. Biomolecules. 2023 Apr 6;13(4):654. doi: 10.3390/biom13040654. Biomolecules. 2023. PMID: 37189401 Free PMC article. - Pectobacterium Phage Jarilo Displays Broad Host Range and Represents a Novel Genus of Bacteriophages Within the Family Autographiviridae.
Pedersen JS, Carstens AB, Djurhuus AM, Kot W, Neve H, Hansen LH. Pedersen JS, et al. Phage (New Rochelle). 2020 Dec 1;1(4):237-244. doi: 10.1089/phage.2020.0037. Epub 2020 Dec 16. Phage (New Rochelle). 2020. PMID: 36147289 Free PMC article. - Diverse Mechanisms of RNA Recombination.
Gmyl AP, Agol VI. Gmyl AP, et al. Mol Biol. 2005;39(4):529-542. doi: 10.1007/s11008-005-0069-x. Mol Biol. 2005. PMID: 32214466 Free PMC article.
References
- Saldanha R, Mohr G, Belfort M, Lambowitz A M. FASEB J. 1993;7:15–24. - PubMed
- Gott J M, Shub D A, Belfort M. Cell. 1986;47:81–87. - PubMed
- Young P, Ohman M, Xu M-Q, Shub D A, Sjöberg B M. J Biol Chem. 1994;269:20229–20232. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources