Cell cycle-regulated histone acetylation required for expression of the yeast HO gene - PubMed (original) (raw)
Cell cycle-regulated histone acetylation required for expression of the yeast HO gene
J E Krebs et al. Genes Dev. 1999.
Abstract
Expression of the yeast HO gene in late G1 of the cell cycle requires the SWI/SNF chromatin remodeling complex, the Gcn5p histone acetyltransferase, and two different sequence-specific transcriptional activators, Swi5p and Swi4p/Swi6p. We have used chromatin immunoprecipitation assays to investigate the role of each of these trans-acting factors in establishing a cell cycle-regulated domain of histone acetylation surrounding the HO upstream regulatory region. We detect a approximately 1-kb domain of H3 and H4 acetylation that is established in mid-G1, prior to and independent of HO transcription, which then declines with kinetics similar to inactivation of HO. This cell cycle burst of histone acetylation requires Gcn5p, SWI/SNF, and the Swi5p activator, but occurs in the absence of the Swi4p activator. We also find that inactivation of the Sin3p/Rpd3p deacetylase complex leads to a high level of acetylation at the HO locus throughout the cell cycle. We propose a sequential model for activation of HO in which the Swi5p-dependent recruitment of the Gcn5p acetyltransferase requires chromatin remodeling events by the SWI/SNF complex.
Figures
Figure 1
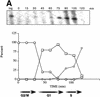
Nucleosomes at the TATA region of HO are subject to _GCN5_-dependent, transient acetylation prior to transcription. (A) Budding index and primer extension analysis of HO transcripts show the cell cycle-dependence of HO expression. Cells were arrested in G2/M by treatment with nocodazole, as described in Materials and Methods. Percentages of cells with no buds (□) (G1), small buds (⋄) (S phase), or large buds (○) (G2/M) were determined at the times shown after release from nocodazole arrest. Samples were taken from these timepoints for either RNA analysis or for ChIPs (see below). HO transcripts are detectable by primer extension in late G1 and peak at the onset of S phase (appearance of small buds; 90–105 min in wild-type cells). (B) Results of ChIP assays for either wild-type or _gcn5_- cells. Chromatin obtained from cells released from nocodazole arrest as described above was immunoprecipitated with antibodies that recognize histone H3 acetylated at positions 9 and 14 (αH3ac.9/14). DNA obtained from either input or immunoprecipitated material was applied to slot blots. Representative slot blots probed for the HO TATA region are shown. (C) Quantitation of representative slot blots. Blots were quantitated with a PhosphorImager; the results are expressed as the ratio of the bound to the input material (IP efficiency). The graph at left shows the results for the HO TATA probe for either GCN5+ or _gcn5_− cells, showing the _GCN5_-dependent peak of acetylation between 60 and 90 min after release from nocodazole arrest. The high levels of acetylation detected in log phase cells, as well as in early S phase, are also partially GCN5 dependent. The middle graph depicts the global levels of _GCN5_-dependent acetylation using the genomic DNA probe, and acetylation detected by the rDNA probe is shown at right. The log- and early S-phase levels of acetylation are high in both total DNA and rDNA, but the late-G1 peak is specific to HO. Data shown are representative of four separate experiments with ChIP extracts from two independent cell synchronizations. (D) ChIP analysis of the wild-type samples from C using antibodies against unacetylated H3. The smaller scale on this graph reflects the weaker IP efficiency of these antibodies. (E) ChIP analysis of wild-type and _gcn5_- cells with antibodies recognizing hyperacetylated histone H4. (Solid bars) Gcn5p; (open bars) gcn5.
Figure 1
Nucleosomes at the TATA region of HO are subject to _GCN5_-dependent, transient acetylation prior to transcription. (A) Budding index and primer extension analysis of HO transcripts show the cell cycle-dependence of HO expression. Cells were arrested in G2/M by treatment with nocodazole, as described in Materials and Methods. Percentages of cells with no buds (□) (G1), small buds (⋄) (S phase), or large buds (○) (G2/M) were determined at the times shown after release from nocodazole arrest. Samples were taken from these timepoints for either RNA analysis or for ChIPs (see below). HO transcripts are detectable by primer extension in late G1 and peak at the onset of S phase (appearance of small buds; 90–105 min in wild-type cells). (B) Results of ChIP assays for either wild-type or _gcn5_- cells. Chromatin obtained from cells released from nocodazole arrest as described above was immunoprecipitated with antibodies that recognize histone H3 acetylated at positions 9 and 14 (αH3ac.9/14). DNA obtained from either input or immunoprecipitated material was applied to slot blots. Representative slot blots probed for the HO TATA region are shown. (C) Quantitation of representative slot blots. Blots were quantitated with a PhosphorImager; the results are expressed as the ratio of the bound to the input material (IP efficiency). The graph at left shows the results for the HO TATA probe for either GCN5+ or _gcn5_− cells, showing the _GCN5_-dependent peak of acetylation between 60 and 90 min after release from nocodazole arrest. The high levels of acetylation detected in log phase cells, as well as in early S phase, are also partially GCN5 dependent. The middle graph depicts the global levels of _GCN5_-dependent acetylation using the genomic DNA probe, and acetylation detected by the rDNA probe is shown at right. The log- and early S-phase levels of acetylation are high in both total DNA and rDNA, but the late-G1 peak is specific to HO. Data shown are representative of four separate experiments with ChIP extracts from two independent cell synchronizations. (D) ChIP analysis of the wild-type samples from C using antibodies against unacetylated H3. The smaller scale on this graph reflects the weaker IP efficiency of these antibodies. (E) ChIP analysis of wild-type and _gcn5_- cells with antibodies recognizing hyperacetylated histone H4. (Solid bars) Gcn5p; (open bars) gcn5.
Figure 1
Nucleosomes at the TATA region of HO are subject to _GCN5_-dependent, transient acetylation prior to transcription. (A) Budding index and primer extension analysis of HO transcripts show the cell cycle-dependence of HO expression. Cells were arrested in G2/M by treatment with nocodazole, as described in Materials and Methods. Percentages of cells with no buds (□) (G1), small buds (⋄) (S phase), or large buds (○) (G2/M) were determined at the times shown after release from nocodazole arrest. Samples were taken from these timepoints for either RNA analysis or for ChIPs (see below). HO transcripts are detectable by primer extension in late G1 and peak at the onset of S phase (appearance of small buds; 90–105 min in wild-type cells). (B) Results of ChIP assays for either wild-type or _gcn5_- cells. Chromatin obtained from cells released from nocodazole arrest as described above was immunoprecipitated with antibodies that recognize histone H3 acetylated at positions 9 and 14 (αH3ac.9/14). DNA obtained from either input or immunoprecipitated material was applied to slot blots. Representative slot blots probed for the HO TATA region are shown. (C) Quantitation of representative slot blots. Blots were quantitated with a PhosphorImager; the results are expressed as the ratio of the bound to the input material (IP efficiency). The graph at left shows the results for the HO TATA probe for either GCN5+ or _gcn5_− cells, showing the _GCN5_-dependent peak of acetylation between 60 and 90 min after release from nocodazole arrest. The high levels of acetylation detected in log phase cells, as well as in early S phase, are also partially GCN5 dependent. The middle graph depicts the global levels of _GCN5_-dependent acetylation using the genomic DNA probe, and acetylation detected by the rDNA probe is shown at right. The log- and early S-phase levels of acetylation are high in both total DNA and rDNA, but the late-G1 peak is specific to HO. Data shown are representative of four separate experiments with ChIP extracts from two independent cell synchronizations. (D) ChIP analysis of the wild-type samples from C using antibodies against unacetylated H3. The smaller scale on this graph reflects the weaker IP efficiency of these antibodies. (E) ChIP analysis of wild-type and _gcn5_- cells with antibodies recognizing hyperacetylated histone H4. (Solid bars) Gcn5p; (open bars) gcn5.
Figure 2
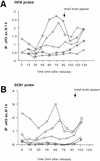
Acetylation at the HO promoter is limited to a 1-kb region encompassing the TATA box and the Swi4p/Swi6p-binding sites. (A) Schematic of the HO promoter showing the probes used in this study. (Hatched bars) TATA; (solid bars) SWI cell cycle box (SCB); (open bars) Swi5p binding site. (B) Results of probe scan of HO for samples precipitated with αH3ac.9/14. The slot blots shown in Fig. 1B were stripped and hybridized with a variety of DNA probes covering the HO promoter and coding region. The results for the TATA probe are the same as shown in Fig. 1C. Probes for the Swi4p/Swi6p-binding sites (SCB1 and SCB2) also detect a peak of acetylation that occurs slightly before the peak detected at the TATA. Probes for either of the Swi5p-binding sites (5A and 5B) or for the HO coding region (5′ ORF or 3′ ORF) do not detect a peak of acetylation. (C) Results of probe scan of HO for samples precipitated with antibodies to hyperacetylated H4. (B,C) (▵) 5′ ORF; (⋄) TATA; (□) SCB1; (○) 5B; (▿) 5A.
Figure 3
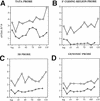
The cell cycle-regulated domain of histone H3 acetylation at HO requires Gcn5p, Swi5p, and an intact SWI/SNF complex. SWI+, _swi2_−, _swi5_−, or _swi4_− cells were synchronized with nocodazole and samples were fixed for chromatin IP after synchronous release. ChIPs were performed with the antibody to diacetylated histone H3 (ac.9/14). The swi4 and swi2 mutants showed a significant delay in recovering from nocodazole arrest and thus synchronies are aligned by the time of bud emergence (arrow at top). Time scale shown on the x axis is from the _SWI_+ synchrony. Data shown are the result of four separate experiments from each strain with two independent cell synchronies. (A) Acetylation at the HO TATA region in wild-type (WT) (○) cells and in swi5 (⊞), swi2 (⋄), swi4 (□), and gcn5 (▵) mutants. (B) Acetylation over the SCB elements in wild-type cells and in swi5, swi2, swi4, and gcn5 mutants.
Figure 4
The entire HO gene is acetylated throughout the cell cycle in sin3Δ cells. _sin3_− cells were synchronized with nocodazole and samples were fixed for chromatin IP after synchronous release. ChIPs were performed with the antibody to diacetylated histone H3 (ac.9/14). In each panel, data from sin3 cells (□) are compared with wild-type (⋄). (A) Acetylation at the HO TATA region in wild-type cells and in sin3 mutants. (B) Acetylation at the 5′ end of the HO coding region in wild-type cells and in sin3 mutants. (C) Acetylation at the upstream Swi5p-binding site in wild-type cells and in sin3 mutants. (D) Genomic H3 acetylation in wild-type and sin3 cells.
Similar articles
- SWI/SNF binding to the HO promoter requires histone acetylation and stimulates TATA-binding protein recruitment.
Mitra D, Parnell EJ, Landon JW, Yu Y, Stillman DJ. Mitra D, et al. Mol Cell Biol. 2006 Jun;26(11):4095-110. doi: 10.1128/MCB.01849-05. Mol Cell Biol. 2006. PMID: 16705163 Free PMC article. - Ordered recruitment of transcription and chromatin remodeling factors to a cell cycle- and developmentally regulated promoter.
Cosma MP, Tanaka T, Nasmyth K. Cosma MP, et al. Cell. 1999 Apr 30;97(3):299-311. doi: 10.1016/s0092-8674(00)80740-0. Cell. 1999. PMID: 10319811 - Global role for chromatin remodeling enzymes in mitotic gene expression.
Krebs JE, Fry CJ, Samuels ML, Peterson CL. Krebs JE, et al. Cell. 2000 Sep 1;102(5):587-98. doi: 10.1016/s0092-8674(00)00081-7. Cell. 2000. PMID: 11007477 - Transcription: gene control by targeted histone acetylation.
Imhof A, Wolffe AP. Imhof A, et al. Curr Biol. 1998 Jun 4;8(12):R422-4. doi: 10.1016/s0960-9822(98)70268-4. Curr Biol. 1998. PMID: 9637914 Review. - Recruitment of chromatin remodelling factors during gene activation via the glucocorticoid receptor N-terminal domain.
Wallberg AE, Flinn EM, Gustafsson JA, Wright AP. Wallberg AE, et al. Biochem Soc Trans. 2000;28(4):410-4. Biochem Soc Trans. 2000. PMID: 10961930 Review.
Cited by
- Distinct roles for the essential MYST family HAT Esa1p in transcriptional silencing.
Clarke AS, Samal E, Pillus L. Clarke AS, et al. Mol Biol Cell. 2006 Apr;17(4):1744-57. doi: 10.1091/mbc.e05-07-0613. Epub 2006 Jan 25. Mol Biol Cell. 2006. PMID: 16436512 Free PMC article. - Inhibition of acetyl coenzyme A carboxylase activity restores expression of the INO1 gene in a snf1 mutant strain of Saccharomyces cerevisiae.
Shirra MK, Patton-Vogt J, Ulrich A, Liuta-Tehlivets O, Kohlwein SD, Henry SA, Arndt KM. Shirra MK, et al. Mol Cell Biol. 2001 Sep;21(17):5710-22. doi: 10.1128/MCB.21.17.5710-5722.2001. Mol Cell Biol. 2001. PMID: 11486011 Free PMC article. - Post-TATA binding protein recruitment clearance of Gcn5-dependent histone acetylation within promoter nucleosomes.
Topalidou I, Papamichos-Chronakis M, Thireos G. Topalidou I, et al. Mol Cell Biol. 2003 Nov;23(21):7809-17. doi: 10.1128/MCB.23.21.7809-7817.2003. Mol Cell Biol. 2003. PMID: 14560024 Free PMC article. - HAT cofactor Trrap regulates the mitotic checkpoint by modulation of Mad1 and Mad2 expression.
Li H, Cuenin C, Murr R, Wang ZQ, Herceg Z. Li H, et al. EMBO J. 2004 Dec 8;23(24):4824-34. doi: 10.1038/sj.emboj.7600479. Epub 2004 Nov 18. EMBO J. 2004. PMID: 15549134 Free PMC article. - Genome-wide identification of Isw2 chromatin-remodeling targets by localization of a catalytically inactive mutant.
Gelbart ME, Bachman N, Delrow J, Boeke JD, Tsukiyama T. Gelbart ME, et al. Genes Dev. 2005 Apr 15;19(8):942-54. doi: 10.1101/gad.1298905. Genes Dev. 2005. PMID: 15833917 Free PMC article.
References
- Amon A, Tyers M, Futcher B, Nasmyth K. Mechanisms that help the yeast cell cycle clock tick: G2 cyclins transcriptionally activate G2 cyclins and repress G1 cyclins. Cell. 1993;74:993–1007. - PubMed
- Bobola N, Jansen RP, Shin TH, Nasmyth K. Asymmetric accumulation of Ash1p in postanaphase nuclei depends on a myosin and restricts yeast mating-type switching to mother cells. Cell. 1996;84:699–709. - PubMed
- Breeden L, Nasmyth K. Cell cycle control of the yeast HO gene: Cis- and trans-acting regulators. Cell. 1987;48:389–397. - PubMed
- Brownell JE, Zhou J, Ranalli T, Kobayashi R, Edmondson DG, Roth SY, Allis CD. Tetrahymena histone acetyltransferase A: A homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell. 1996;84:843–851. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases