Mapping of functional elements in the stem-anchor region of tick-borne encephalitis virus envelope protein E - PubMed (original) (raw)
Mapping of functional elements in the stem-anchor region of tick-borne encephalitis virus envelope protein E
S L Allison et al. J Virol. 1999 Jul.
Abstract
Envelope protein E of the flavivirus tick-borne encephalitis virus mediates membrane fusion, and the structure of the N-terminal 80% of this 496-amino-acid-long protein has been shown to differ significantly from that of other viral fusion proteins. The structure of the carboxy-terminal 20%, the stem-anchor region, is not known. It contains sequences that are important for membrane anchoring, interactions with prM (the precursor of membrane protein M) during virion assembly, and low-pH-induced structural changes associated with the fusion process. To identify specific functional elements in this region, a series of C-terminal deletion mutants were constructed and the properties of the resulting truncated recombinant E proteins were examined. Full-length E proteins and proteins lacking the second of two predicted transmembrane segments were secreted in a particulate form when coexpressed with prM, whereas deletion of both segments resulted in the secretion of soluble homodimeric E proteins. Sites located within a predicted alpha-helical region of the stem (amino acids 431 to 449) and the first membrane-spanning region (amino acids 450 to 472) were found to be important for the stability of the prM-E heterodimer but not essential for prM-mediated intracellular transport and secretion of soluble E proteins. A separate site in the stem, also corresponding to a predicted alpha-helix (amino acids 401 to 413), was essential for the conversion of soluble protein E dimers to a homotrimeric form upon low-pH treatment, a process resembling the transition to the fusogenic state in whole virions. This functional mapping will aid in the understanding of the molecular mechanisms of membrane fusion and virus assembly.
Figures
FIG. 1
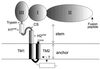
Schematic diagram of a TBE virus protein E monomer (not to scale) which, in the native structure, forms a homodimer with a head-to-tail orientation. The three domains of the sE fragment (I, II, and III), which was cleaved from purified virions by trypsin and used previously for structure determination (32), are represented by shaded ovals, and the positions of the putative fusion peptide at the tip of domain II and the trypsin cleavage site next to domain III are indicated. The part below the trypsin cleavage site represents the C-terminal 20% of the E protein, for which only sequence-based structure predictions are available (36). The viral membrane is represented by parallel lines, and the portions of E corresponding to the stem and anchor regions are indicated. H1pred (residues 401 to 413) and H2pred (residues 431 to 449) indicate predicted α-helical regions in the stem, the CS element (residues 414 to 430) separates these putative helices, and TM1 (residues 450 to 471) and TM2 (residues 473 to 496) are predicted transmembrane segments (31). H2pred is contiguous with TM1, and these together may constitute a continuous α-helix with its C-terminal half spanning the viral membrane. TM2, which normally serves as the signal sequence for nonstructural protein NS1, is depicted as traversing the membrane, but it is not known whether it actually remains in the membrane after cleavage by signalase, which separates the E and NS1 proteins during processing of the viral polyprotein (33).
FIG. 2
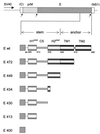
C-terminal truncations in recombinant E proteins. The diagram shows the amino acid (aa) positions of the C-terminal truncations and which of the predicted structural elements of the stem-anchor region were present in each construct (Fig. 1 contains the definitions of the abbreviations). These proteins are designated by an E followed by the number of the last amino acid residue. The portion of the TBE genome containing the prM and E genes, cloned into a plasmid vector under the control of the SV40 early promoter (1), is shown at the top. Small arrows indicate the sites where the polyprotein is cleaved by signalase (33), and the short flanking sequences belonging to the C and NS1 coding regions are labeled.
FIG. 3
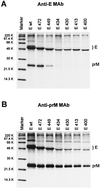
SDS-PAGE of recombinant TBE virus proteins after immunoprecipitation with E-specific (A) and prM-specific (B) MAbs. COS cells were transfected with recombinant plasmids encoding prM together with one of the truncated forms of protein E shown in Fig. 2, radiolabeled, and lysed in a buffer containing 1% Triton X-100. The proteins were then immunoprecipitated, separated by SDS-PAGE, and visualized by fluorography. The values on the left are molecular weights.
FIG. 4
Cosedimentation analysis of prM and truncated E proteins. The cell lysates used in the experiment depicted in Fig. 3 were analyzed by centrifugation in sucrose gradients (5 to 20%, wt/wt) containing 0.5% Triton X-100. Proteins were immunoprecipitated from the gradient fractions by using a polyclonal antiserum recognizing both prM and protein E, separated by SDS-PAGE, and visualized by fluorography. Panels: A, prM and Ewt; B, prM and E472; C, prM and E449; D, prM and E430. The sedimentation direction is left to right.
FIG. 5
Pelleting efficiencies and HA activities of secreted E proteins. Cleared supernatants from transfected COS cells were subjected to ultracentrifugation, and the amounts of protein E in the pellet and supernatant fractions after centrifugation were quantitated by ELISA. HA activity was determined both before and after pelleting. +, HA activity detected both before pelleting and in the pelleted fraction; −, no HA activity detected.
FIG. 6
Cross-linking analysis of soluble truncated E proteins. Partially purified E proteins were analyzed by SDS-PAGE and immunoblotting after treatment with the cross-linking reagent DMS. The positions of the protein E monomer and dimer bands are indicated. +, cross-linker added; −, no cross-linker.
FIG. 7
Effect of coexpression with prM on E400 secretion efficiency. COS cells were transfected with expression plasmids encoding prM and E400 in the same construct (A), E400 alone (B), or prM and E400 on separate plasmids (C). At 29 h posttransfection, a protein E-specific four-layer ELISA (20) was used to detect secreted protein E in the supernatant. The transfection efficiency, as judged by immunofluorescence using a protein E-specific MAb, was the same (∼50%) in all three samples (data not shown).
FIG. 8
Sedimentation analysis of detergent-solubilized E proteins at pH 8.0 (solid curves) or after pretreatment at pH 6.0 and back-neutralization to pH 8.0 (dashed curves). Samples were centrifuged in sucrose gradients (7 to 20%, wt/wt) containing 0.1% Triton X-100, and the amount of protein E in each fraction was quantitated by ELISA. For the insets, material from the peak fractions was subjected to DMS cross-linking and immunoblotting as described in the legend to Fig. 6. M, monomer; D, dimer; T, trimer.
Similar articles
- Structural requirements for low-pH-induced rearrangements in the envelope glycoprotein of tick-borne encephalitis virus.
Stiasny K, Allison SL, Marchler-Bauer A, Kunz C, Heinz FX. Stiasny K, et al. J Virol. 1996 Nov;70(11):8142-7. doi: 10.1128/JVI.70.11.8142-8147.1996. J Virol. 1996. PMID: 8892942 Free PMC article. - Recombinant subviral particles from tick-borne encephalitis virus are fusogenic and provide a model system for studying flavivirus envelope glycoprotein functions.
Schalich J, Allison SL, Stiasny K, Mandl CW, Kunz C, Heinz FX. Schalich J, et al. J Virol. 1996 Jul;70(7):4549-57. doi: 10.1128/JVI.70.7.4549-4557.1996. J Virol. 1996. PMID: 8676481 Free PMC article. - Membrane interactions of the tick-borne encephalitis virus fusion protein E at low pH.
Stiasny K, Allison SL, Schalich J, Heinz FX. Stiasny K, et al. J Virol. 2002 Apr;76(8):3784-90. doi: 10.1128/jvi.76.8.3784-3790.2002. J Virol. 2002. PMID: 11907218 Free PMC article. - Molecular mechanisms of flavivirus membrane fusion.
Stiasny K, Fritz R, Pangerl K, Heinz FX. Stiasny K, et al. Amino Acids. 2011 Nov;41(5):1159-63. doi: 10.1007/s00726-009-0370-4. Epub 2009 Nov 1. Amino Acids. 2011. PMID: 19882217 Review. - The molecular biology of tick-borne encephalitis virus. Review article.
Heinz FX, Mandl CW. Heinz FX, et al. APMIS. 1993 Oct;101(10):735-45. doi: 10.1111/j.1699-0463.1993.tb00174.x. APMIS. 1993. PMID: 8267950 Review.
Cited by
- Mechanism of membrane fusion by viral envelope proteins.
Harrison SC. Harrison SC. Adv Virus Res. 2005;64:231-61. doi: 10.1016/S0065-3527(05)64007-9. Adv Virus Res. 2005. PMID: 16139596 Free PMC article. - Early Events in Japanese Encephalitis Virus Infection: Viral Entry.
Yun SI, Lee YM. Yun SI, et al. Pathogens. 2018 Aug 13;7(3):68. doi: 10.3390/pathogens7030068. Pathogens. 2018. PMID: 30104482 Free PMC article. Review. - Identification of Potential Lead Molecules for Zika Envelope Protein from In Silico Perspective.
Chellasamy SK, Devarajan S. Chellasamy SK, et al. Avicenna J Med Biotechnol. 2019 Jan-Mar;11(1):94-103. Avicenna J Med Biotechnol. 2019. PMID: 30800249 Free PMC article. - Neuroadapted yellow fever virus strain 17D: a charged locus in domain III of the E protein governs heparin binding activity and neuroinvasiveness in the SCID mouse model.
Nickells J, Cannella M, Droll DA, Liang Y, Wold WS, Chambers TJ. Nickells J, et al. J Virol. 2008 Dec;82(24):12510-9. doi: 10.1128/JVI.00458-08. Epub 2008 Oct 8. J Virol. 2008. PMID: 18842715 Free PMC article.
References
- Allison S L, Mandl C W, Kunz C, Heinz F X. Expression of cloned envelope protein genes from the flavivirus tick-borne encephalitis virus in mammalian cells and random mutagenesis by PCR. Virus Genes. 1994;8:187–198. - PubMed
- Blacklow S C, Lu M, Kim P S. A trimeric subdomain of the simian immunodeficiency virus envelope glycoprotein. Biochemistry. 1995;34:14955–14962. - PubMed
- Bullough P A, Hughson F M, Skehel J J, Wiley D C. Structure of influenza haemagglutinin at the pH of membrane fusion. Nature. 1994;371:37–43. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources