Colocalization and membrane association of murine hepatitis virus gene 1 products and De novo-synthesized viral RNA in infected cells - PubMed (original) (raw)
Colocalization and membrane association of murine hepatitis virus gene 1 products and De novo-synthesized viral RNA in infected cells
S T Shi et al. J Virol. 1999 Jul.
Abstract
Murine hepatitis virus (MHV) gene 1, the 22-kb polymerase (pol) gene, is first translated into a polyprotein and subsequently processed into multiple proteins by viral autoproteases. Genetic complementation analyses suggest that the majority of the gene 1 products are required for viral RNA synthesis. However, there is no physical evidence supporting the association of any of these products with viral RNA synthesis. We have now performed immunofluorescent-staining studies with four polyclonal antisera to localize various MHV-A59 gene 1 products in virus-infected cells. Immunoprecipitation experiments showed that these antisera detected proteins representing the two papain-like proteases and the 3C-like protease encoded by open reading frame (ORF) 1a, the putative polymerase (p100) and a p35 encoded by ORF 1b, and their precursors. De novo-synthesized viral RNA was labeled with bromouridine triphosphate in lysolecithin-permeabilized MHV-infected cells. Confocal microscopy revealed that all of the viral proteins detected by these antisera colocalized with newly synthesized viral RNA in the cytoplasm, particularly in the perinuclear region of infected cells. Several cysteine and serine protease inhibitors, i.e., E64d, leupeptin, and zinc chloride, inhibited viral RNA synthesis without affecting the localization of viral proteins, suggesting that the processing of the MHV gene 1 polyprotein is tightly associated with viral RNA synthesis. Dual labeling with antibodies specific for cytoplasmic membrane structures showed that MHV gene 1 products and RNA colocalized with the Golgi apparatus in HeLa cells. However, in murine 17CL-1 cells, the viral proteins and viral RNA did not colocalize with the Golgi apparatus but, instead, partially colocalized with the endoplasmic reticulum. Our results provide clear physical evidence that several MHV gene 1 products, including the proteases and the polymerase, are associated with the viral RNA replication-transcription machinery, which may localize to different membrane structures in different cell lines.
Figures
FIG. 1
Domains and processed products of the MHV gene 1 polyprotein. Predicted MHV functional domains indicated here were identified by amino acid sequence homology to known viral functional domains (22, 29). Abbreviations: PCP, papain-like cysteine protease; 3Cpro, poliovirus 3C-like protease; MP, membrane-associated protein; HEL, helicase; POL, polymerase. Regions used to generate fusion proteins and ultimately rabbit polyclonal antisera are indicated under the amino acid scale. The processing of ORF 1a products depicted here is based on previous findings (2, 6, 12, 13, 20, 33, 34, 45, 48) and results shown in Fig. 5 and is slightly different from that reported by Denison et al. (15). The dashed lines represent products of uncertain nature.
FIG. 2
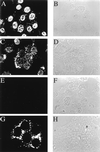
Detection of de novo-synthesized viral RNA by BrUTP labeling. MHV-A59-infected 17CL-1 cells were permeabilized with lysolecithin, treated with BrUTP at 8.5 h postinfection, and fixed 10 min later. The cells were stained with a monoclonal antibody against BrdU. Representative images of BrUTP labeling are shown for mock-infected (A and E) or MHV-infected (C and G) cells in the presence (E and G) or absence (A and C) of 5 μg of actinomycin D per ml. Actinomycin D was added 1 h before BrUTP labeling. B, D, F, and H are the phase-contrast images of A, C, E, and G, respectively.
FIG. 3
Colocalization of MHV gene 1 products with de novo-synthesized viral RNA. MHV-A59-infected cells were permeabilized and labeled with BrUTP at 8.5 h (17CL-1 cells) or 7 h (HeLa-MHVR cells) postinfection. Actinomycin D (5 μg/ml) was added 1 h before BrUTP labeling. The cells were then double stained with rabbit polyclonal antisera against D3, D12, POL, or D23 regions of gene 1 polyprotein and the mouse monoclonal anti-BrdU antibody (RNA). (A) 17CL-1 cells; (B) HeLa-MHVR cells.
FIG. 4
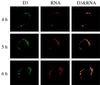
Kinetic study of MHV RNA synthesis. MHV-A59-infected 17CL-1 cells were permeabilized and subjected to BrUTP labeling at 4, 5, and 6 h postinfection in the presence of 5 μg of actinomycin D per ml. Actinomycin D was added 1 h before BrUTP labeling. The cells were then double stained with the D3 antiserum and mouse monoclonal BrdU antibody (RNA) as described in the Materials and Methods.
FIG. 5
(A) Immunoprecipitation of MHV ORF 1b proteins by anti-D18 and anti-D23. HeLa-MHVR cells were infected with MHV-A59 and treated with 2 μg of actinomycin D per ml at 1 h postinfection for the duration of the experiment. At 2.5 h postinfection, the medium was replaced for 30 min with DMEM lacking methionine. Proteins were metabolically radiolabeled with [35S]methionine for 2 h, followed by a 30-min chase period in complete medium. Cell lysates were prepared at 5.5 h postinfection and subjected to immunoprecipitation as described previously (45). (B) Inhibition of the processing of MHV gene 1 polyprotein by E64d. E64d (400 μg/ml) was added to the cells at 2 h postinfection and maintained in the medium until the cell lysates were prepared at 5.5 h postinfection. The immunoprecipitated viral proteins are indicated by arrows.
FIG. 6
Inhibition of MHV RNA synthesis by protein synthesis and protease inhibitors. MHV-A59-infected cells were untreated (A and D) or treated with 10 μg of cycloheximide per ml (B and E) or 400 μg of E64d per ml (C and F) 1.5 h before permeabilization and BrUTP labeling. The cells were labeled with BrUTP for 40 min and double stained with the D3 (A to C) and BrdU (RNA) (D to F) antibodies.
FIG. 7
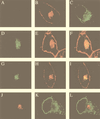
Colocalization of MHV RNA and gene 1 products with the Golgi apparatus in HeLa-MHVR cells. (A) Mock-infected cells were stained with the D3 antiserum; (B and C) mock-infected cells were double stained with WGA (Golgi) (B) and the GRP78 antibody (ER) (C); (D to F) MHV-A59-infected cells were permeabilized and labeled with BrUTP, followed by double staining with the BrdU antibody (RNA) (D) and WGA (E); (G to I) virus-infected cells were double stained with the D3 antiserum (G) and WGA (H); (J to L) virus-infected cells were double stained with antibodies against D3 (J) and GRP78 (ER) (K). Panels F, I, and L are the superimposed images of D and E, G and H, and J and K, respectively.
FIG. 8
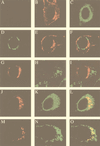
Partial colocalization of MHV gene 1 products with the ER in 17CL-1 cells. (A) Mock-infected cells were stained with the D3 antiserum; (B and C) mock-infected cells were double stained with WGA (Golgi) (B) and the GRP78 antibody (ER) (C); (D to F) MHV-A59-infected cells were double stained with the D3 antiserum (D) and WGA (Golgi) (E); (G to I) virus-infected cells were double stained with antibodies against D3 (G) and LAMP-1 (lysosome) (H); (J to L) virus-infected cells were double stained with antibodies against D3 (J) and GRP78 (ER) (K); (M to O) MHV-A59-infected cells were permeabilized and labeled with BrUTP, followed by double staining with antibodies against BrdU (RNA) (M) and GRP78 (ER) (N). Panels F, I, L, and O are the superimposed images of D and E, G and H, J and K, and M and N, respectively.
Similar articles
- The putative helicase of the coronavirus mouse hepatitis virus is processed from the replicase gene polyprotein and localizes in complexes that are active in viral RNA synthesis.
Denison MR, Spaan WJ, van der Meer Y, Gibson CA, Sims AC, Prentice E, Lu XT. Denison MR, et al. J Virol. 1999 Aug;73(8):6862-71. doi: 10.1128/JVI.73.8.6862-6871.1999. J Virol. 1999. PMID: 10400784 Free PMC article. - Identification of mouse hepatitis virus papain-like proteinase 2 activity.
Kanjanahaluethai A, Baker SC. Kanjanahaluethai A, et al. J Virol. 2000 Sep;74(17):7911-21. doi: 10.1128/jvi.74.17.7911-7921.2000. J Virol. 2000. PMID: 10933699 Free PMC article. - Coronavirus protein processing and RNA synthesis is inhibited by the cysteine proteinase inhibitor E64d.
Kim JC, Spence RA, Currier PF, Lu X, Denison MR. Kim JC, et al. Virology. 1995 Apr 1;208(1):1-8. doi: 10.1006/viro.1995.1123. Virology. 1995. PMID: 11831690 Free PMC article. - RNA replication of mouse hepatitis virus takes place at double-membrane vesicles.
Gosert R, Kanjanahaluethai A, Egger D, Bienz K, Baker SC. Gosert R, et al. J Virol. 2002 Apr;76(8):3697-708. doi: 10.1128/jvi.76.8.3697-3708.2002. J Virol. 2002. PMID: 11907209 Free PMC article. - Functional and genetic analysis of coronavirus replicase-transcriptase proteins.
Sawicki SG, Sawicki DL, Younker D, Meyer Y, Thiel V, Stokes H, Siddell SG. Sawicki SG, et al. PLoS Pathog. 2005 Dec;1(4):e39. doi: 10.1371/journal.ppat.0010039. Epub 2005 Dec 9. PLoS Pathog. 2005. PMID: 16341254 Free PMC article. Review.
Cited by
- SARS-CoV-2 nsp15 preferentially degrades AU-rich dsRNA via its dsRNA nickase activity.
Wang X, Zhu B. Wang X, et al. Nucleic Acids Res. 2024 May 22;52(9):5257-5272. doi: 10.1093/nar/gkae290. Nucleic Acids Res. 2024. PMID: 38634805 Free PMC article. - Structural basis for polyuridine tract recognition by SARS-CoV-2 Nsp15.
Ito F, Yang H, Zhou ZH, Chen XS. Ito F, et al. bioRxiv [Preprint]. 2023 Nov 20:2023.11.17.567629. doi: 10.1101/2023.11.17.567629. bioRxiv. 2023. PMID: 38045375 Free PMC article. Updated. Preprint. - Regulation of coronavirus nsp15 cleavage specificity by RNA structure.
Salukhe I, Choi R, Van Voorhis W, Barrett L, Hyde J. Salukhe I, et al. PLoS One. 2023 Aug 24;18(8):e0290675. doi: 10.1371/journal.pone.0290675. eCollection 2023. PLoS One. 2023. PMID: 37616296 Free PMC article. - Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Godoy AS, Nakamura AM, Douangamath A, Song Y, Noske GD, Gawriljuk VO, Fernandes RS, Pereira HDM, Oliveira KIZ, Fearon D, Dias A, Krojer T, Fairhead M, Powell A, Dunnet L, Brandao-Neto J, Skyner R, Chalk R, Bajusz D, Bege M, Borbás A, Keserű GM, von Delft F, Oliva G. Godoy AS, et al. Nucleic Acids Res. 2023 Jun 9;51(10):5255-5270. doi: 10.1093/nar/gkad314. Nucleic Acids Res. 2023. PMID: 37115000 Free PMC article. - Porcine Epidemic Diarrhea Virus Infection Induces Autophagosome Formation but Inhibits Autolysosome Formation during Replication.
Park JY, Ryu J, Hong EJ, Shin HJ. Park JY, et al. Viruses. 2022 May 15;14(5):1050. doi: 10.3390/v14051050. Viruses. 2022. PMID: 35632790 Free PMC article.
References
- Bi W, Pinon J D, Hughes S, Bonilla P J, Holmes K V, Weiss S R, Leibowitz J L. Localization of mouse hepatitis virus open reading frame 1A derived proteins. J Neurovirol. 1998;4:594–605. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials