Molecular subtyping of Clostridium perfringens by pulsed-field gel electrophoresis to facilitate food-borne-disease outbreak investigations - PubMed (original) (raw)
Molecular subtyping of Clostridium perfringens by pulsed-field gel electrophoresis to facilitate food-borne-disease outbreak investigations
S E Maslanka et al. J Clin Microbiol. 1999 Jul.
Abstract
Clostridium perfringens is a common cause of food-borne illness. The illness is characterized by profuse diarrhea and acute abdominal pain. Since the illness is usually self-limiting, many cases are undiagnosed and/or not reported. Investigations are often pursued after an outbreak involving large numbers of people in institutions, at restaurants, or at catered meals. Serotyping has been used in the past to assist epidemiologic investigations of C. perfringens outbreaks. However, serotyping reagents are not widely available, and many isolates are often untypeable with existing reagents. We developed a pulsed-field gel electrophoresis (PFGE) method for molecular subtyping of C. perfringens isolates to aid in epidemiologic investigations of food-borne outbreaks. Six restriction endonucleases (SmaI, ApaI, FspI, MluI, KspI, and XbaI) were evaluated with a select panel of C. perfringens strains. SmaI was chosen for further studies because it produced 11 to 13 well-distributed bands of 40 to approximately 1,100 kb which provided good discrimination between isolates. Seventeen distinct patterns were obtained with 62 isolates from seven outbreak investigations or control strains. In general, multiple isolates from a single individual had indistinguishable PFGE patterns. Epidemiologically unrelated isolates (outbreak or control strains) had unique patterns; isolates from different individuals within an outbreak had similar, if not identical, patterns. PFGE identifies clonal relationships of isolates which will assist epidemiologic investigations of food-borne-disease outbreaks caused by C. perfringens.
Figures
FIG. 1
Dendrogram of PFGE subtype patterns of all C. perfringens control strains and outbreak isolates. Analysis range was 40 to 1,400 kb. CPerf1 was used as the reference standard.
FIG. 2
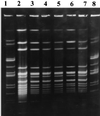
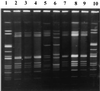
PFGE of C. perfringens strains associated with a single outbreak (NJ 1997). Lanes 1 and 8, reference standard Cperf1; lanes 2 through 7, one isolate from each individual in the outbreak (subtype pattern L).
FIG. 3
PFGE of C. perfringens associated with a single outbreak with possible multiple infecting strains (CA 1983). Lanes 1 and 10, reference standard Cperf1; lanes 2 through 4 and 7 through 9, individuals with subtype pattern A; lane 5, individual with subtype pattern G; lane 6, individual with subtype pattern N.
Similar articles
- Molecular typing of Clostridium perfringens from a food-borne disease outbreak in a nursing home: ribotyping versus pulsed-field gel electrophoresis.
Schalch B, Bader L, Schau HP, Bergmann R, Rometsch A, Maydl G, Kessler S. Schalch B, et al. J Clin Microbiol. 2003 Feb;41(2):892-5. doi: 10.1128/JCM.41.2.892-895.2003. J Clin Microbiol. 2003. PMID: 12574310 Free PMC article. - Molecular epidemiology of Clostridium perfringens related to food-borne outbreaks of disease in Finland from 1984 to 1999.
Lukinmaa S, Takkunen E, Siitonen A. Lukinmaa S, et al. Appl Environ Microbiol. 2002 Aug;68(8):3744-9. doi: 10.1128/AEM.68.8.3744-3749.2002. Appl Environ Microbiol. 2002. PMID: 12147468 Free PMC article. - Prevalence of the enterotoxin gene and clonality of Clostridium perfringens strains associated with food-poisoning outbreaks.
Ridell J, Björkroth J, Eisgrüber H, Schalch B, Stolle A, Korkeala H. Ridell J, et al. J Food Prot. 1998 Feb;61(2):240-3. doi: 10.4315/0362-028x-61.2.240. J Food Prot. 1998. PMID: 9708289 - Diversity of Clostridium perfringens isolates from various sources and prevalence of conjugative plasmids.
Park M, Deck J, Foley SL, Nayak R, Songer JG, Seibel JR, Khan SA, Rooney AP, Hecht DW, Rafii F. Park M, et al. Anaerobe. 2016 Apr;38:25-35. doi: 10.1016/j.anaerobe.2015.11.003. Epub 2015 Nov 30. Anaerobe. 2016. PMID: 26608548 - [Clostridium perfringens].
Komatsu H, Inui A, Sogo T, Fujisawa T. Komatsu H, et al. Nihon Rinsho. 2012 Aug;70(8):1357-61. Nihon Rinsho. 2012. PMID: 22894072 Review. Japanese.
Cited by
- Molecular typing of Clostridium perfringens from a food-borne disease outbreak in a nursing home: ribotyping versus pulsed-field gel electrophoresis.
Schalch B, Bader L, Schau HP, Bergmann R, Rometsch A, Maydl G, Kessler S. Schalch B, et al. J Clin Microbiol. 2003 Feb;41(2):892-5. doi: 10.1128/JCM.41.2.892-895.2003. J Clin Microbiol. 2003. PMID: 12574310 Free PMC article. - Enterotoxigenicity and genetic relatedness of Clostridium perfringens isolates from retail foods in the United States.
Lin YT, Labbe R. Lin YT, et al. Appl Environ Microbiol. 2003 Mar;69(3):1642-6. doi: 10.1128/AEM.69.3.1642-1646.2003. Appl Environ Microbiol. 2003. PMID: 12620854 Free PMC article. - Prevalence of IncFIB Plasmids Found among Salmonella enterica Serovar Schwarzengrund Isolates from Animal Sources in Taiwan Using Whole-Genome Sequencing.
Li IC, Wu HH, Chen ZW, Chou CH. Li IC, et al. Pathogens. 2021 Aug 13;10(8):1024. doi: 10.3390/pathogens10081024. Pathogens. 2021. PMID: 34451486 Free PMC article. - Evidence for Infections by the Same Strain of Beta 2-toxigenic Clostridium perfringens Type A Acquired in One Hospital Ward.
Salamon D, Ochońska D, Wojak I, Mikołajczyk E, Bulanda M, Brzychczy-Włoch M. Salamon D, et al. Pol J Microbiol. 2019 Sep;68(3):323-329. doi: 10.33073/pjm-2019-035. Epub 2019 Sep 3. Pol J Microbiol. 2019. PMID: 31880878 Free PMC article. - Comparison of DNA fingerprinting methods for use in investigation of type E botulism outbreaks in the Canadian Arctic.
Leclair D, Pagotto F, Farber JM, Cadieux B, Austin JW. Leclair D, et al. J Clin Microbiol. 2006 May;44(5):1635-44. doi: 10.1128/JCM.44.5.1635-1644.2006. J Clin Microbiol. 2006. PMID: 16672387 Free PMC article.
References
- Bean N H, Goulding J S, Lao C, Angulo F J. Surveillance for foodborne-disease outbreaks—United States, 1988–1992. CDC surveillance summaries, 25 October 1996. Morbid Mortal Weekly Rep. 1996;45(SS-5):1–67. - PubMed
- Bean N H, Griffin P M. Foodborne disease outbreaks in the United States, 1973–1987: pathogens, vehicles, and trends. J Food Prot. 1990;53:804–817. - PubMed
- Brett M M, Gilbert R J. 1525 outbreaks of Clostridium perfringens food poisoning, 1970–1996. Rev Med Microbiol. 1997;8(Suppl. 1):S64–S65.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous