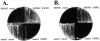A role for ubiquitination in mitochondrial inheritance in Saccharomyces cerevisiae - PubMed (original) (raw)
A role for ubiquitination in mitochondrial inheritance in Saccharomyces cerevisiae
H A Fisk et al. J Cell Biol. 1999.
Abstract
The smm1 mutation suppresses defects in mitochondrial distribution and morphology caused by the mdm1-252 mutation in the yeast Saccharomyces cerevisiae. Cells harboring only the smm1 mutation themselves display temperature-sensitive growth and aberrant mitochondrial inheritance and morphology at the nonpermissive temperature. smm1 maps to RSP5, a gene encoding an essential ubiquitin-protein ligase. The smm1 defects are suppressed by overexpression of wild-type ubiquitin but not by overexpression of mutant ubiquitin in which lysine-63 is replaced by arginine. Furthermore, overexpression of this mutant ubiquitin perturbs mitochondrial distribution and morphology in wild-type cells. Site-directed mutagenesis revealed that the ubiquitin ligase activity of Rsp5p is essential for its function in mitochondrial inheritance. A second mutation, smm2, which also suppressed mdm1-252 defects, but did not cause aberrant mitochondrial distribution and morphology, mapped to BUL1, encoding a protein interacting with Rsp5p. These results indicate that protein ubiquitination mediated by Rsp5p plays an essential role in mitochondrial inheritance, and reveal a novel function for protein ubiquitination.
Figures
Figure 1
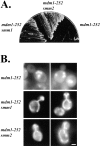
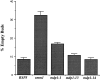
Suppression of mdm1-252 defects by smm1 and smm2. (A) Cells harboring the mdm1-252 mutation (MYY721), or mdm1-252 together with the smm1 (MYY803) or smm2 (MYY804) mutation were cultured on YPD medium for 48 h at 37°C. (B) Strains MYY721, MYY803, and MYY804 were grown on YPD at 23°C, incubated for 4 h at 37°C, and mitochondria were visualized by staining with DASPMI and fluorescence microscopy. Shown are two representative cells for each strain. Bar, 2 μm.
Figure 2
smm1 and smm2 confer temperature-sensitive growth. Strains MYY803 and MYY804 were backcrossed to MYY291 to separate mdm1-252 from smm1 and smm2, respectively. Spores of the indicated genotypes were plated on YPD medium and incubated 48 h at 37°C. (A) Growth at 37°C of spores from a cross of MYY803 and MYY291; (B) growth at 37°C of spores from a cross of MYY804 and MYY291.
Figure 3
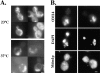
The smm1 mutation causes mitochondrial inheritance defects. Strain MYY809 (smm1) was cultured on YPD medium at 23°C and then incubated for 4 h at 23°C or 37°C. (A) Cells were stained with DASPMI to visualize mitochondria. Bar, 2 μm. (B) Cells which had been shifted to 37°C for 4 h were fixed with formaldehyde and processed for indirect immunofluorescence. Mitochondria were detected with anti-OM14 followed by fluorescein-conjugated donkey anti–mouse IgG (top). Mitochondrial and nuclear DNAs were stained with DAPI (middle). Mdm1p was detected with affinity-purified anti-Mdm1p followed by rhodamine-conjugated donkey anti– rabbit IgG (bottom). Shown are two representative cells. Bar, 2 μm.
Figure 4
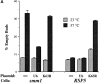
Ubiquitin overexpression affects mitochondrial distribution and morphology. smm1 mutant cells (MYY823) and wild-type cells (MYY290) which harbored no plasmid (−), plasmid pUb (Ub), or plasmid pTER23 (K63R) were cultured at 23°C on minimal medium lacking uracil. CuSO4 was added to 100 μM, and cells were incubated for 2 h at 23°C or 37°C. (A) Cells were fixed, stained with DAPI, and examined by fluorescence microscopy. Budded cells were scored as possessing empty buds if no mitochondrial DNA nucleoids were observed in the bud. Percentages of empty buds were determined for duplicate samples with at least 300 budded cells counted per sample. (B) Wild-type cells (MYY290) transformed with pUb (Ub) or pTER23 (Ub K63R) were incubated at 37°C for 2 h in the presence of 100 μm CuSO4 and stained with DASPMI. Cells were examined by fluorescence (DASPMI) or phase-contrast (Phase) microscopy. Bar, 2 μm.
Figure 4
Ubiquitin overexpression affects mitochondrial distribution and morphology. smm1 mutant cells (MYY823) and wild-type cells (MYY290) which harbored no plasmid (−), plasmid pUb (Ub), or plasmid pTER23 (K63R) were cultured at 23°C on minimal medium lacking uracil. CuSO4 was added to 100 μM, and cells were incubated for 2 h at 23°C or 37°C. (A) Cells were fixed, stained with DAPI, and examined by fluorescence microscopy. Budded cells were scored as possessing empty buds if no mitochondrial DNA nucleoids were observed in the bud. Percentages of empty buds were determined for duplicate samples with at least 300 budded cells counted per sample. (B) Wild-type cells (MYY290) transformed with pUb (Ub) or pTER23 (Ub K63R) were incubated at 37°C for 2 h in the presence of 100 μm CuSO4 and stained with DASPMI. Cells were examined by fluorescence (DASPMI) or phase-contrast (Phase) microscopy. Bar, 2 μm.
Figure 5
Differential effects of rsp5 mutations on mitochondrial inheritance. Haploid _rsp5_-null strains harboring plasmids containing either wild-type RSP5 (MYY826) or mutant forms of rsp5 with the smm1 (MYY829), mdp1-1 (MYY832), mdp1-13 (MYY833), or mdp1-14 (MYY834) mutation were cultured on minimal medium lacking uracil at 23°C. Cells were incubated at 37°C for 2 h, stained with DASPMI, and examined by fluorescence microscopy. Percentages of empty buds were determined for budded cells in duplicate samples with at least 200 cells counted per sample.
Figure 6
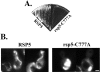
Ubiquitin ligase activity of Rsp5p is required for mitochondrial inheritance. Cells harboring the smm1 mutation (MYY823) were transformed with centromere-based plasmids encoding wild-type RSP5 (pRS316-RSP5) or rsp5 bearing the C777A mutation (pRS316-CA). (A) Transformants were incubated on YPD medium at 37°C for 48 h. (B) Cells were cultured in minimal medium lacking uracil, shifted to 37°C for 4 h, stained with DASPMI, and examined by fluorescence microscopy. Bar, 2 μm.
Figure 7
Mutations in ubiquitin-conjugating enzymes cause defects in mitochondrial morphology and inheritance. Cells harboring null mutations in both ubc4 and ubc5 (LHY21) were cultured at 23°C on YPD medium, incubated at 37°C for 4 h, stained with DASPMI, and examined by fluorescence microscopy. Arrows indicate daughter buds without mitochondria. Bar, 2 μm.
Similar articles
- Bul1, a new protein that binds to the Rsp5 ubiquitin ligase in Saccharomyces cerevisiae.
Yashiroda H, Oguchi T, Yasuda Y, Toh-E A, Kikuchi Y. Yashiroda H, et al. Mol Cell Biol. 1996 Jul;16(7):3255-63. doi: 10.1128/MCB.16.7.3255. Mol Cell Biol. 1996. PMID: 8668140 Free PMC article. - Rsp5p, a new link between the actin cytoskeleton and endocytosis in the yeast Saccharomyces cerevisiae.
Kamińska J, Gajewska B, Hopper AK, Zoładek T. Kamińska J, et al. Mol Cell Biol. 2002 Oct;22(20):6946-8. doi: 10.1128/MCB.22.20.6946-6958.2002. Mol Cell Biol. 2002. PMID: 12242276 Free PMC article. - Ubiquitin metabolism affects cellular response to volatile anesthetics in yeast.
Wolfe D, Reiner T, Keeley JL, Pizzini M, Keil RL. Wolfe D, et al. Mol Cell Biol. 1999 Dec;19(12):8254-62. doi: 10.1128/MCB.19.12.8254. Mol Cell Biol. 1999. PMID: 10567550 Free PMC article. - Versatile role of the yeast ubiquitin ligase Rsp5p in intracellular trafficking.
Belgareh-Touzé N, Léon S, Erpapazoglou Z, Stawiecka-Mirota M, Urban-Grimal D, Haguenauer-Tsapis R. Belgareh-Touzé N, et al. Biochem Soc Trans. 2008 Oct;36(Pt 5):791-6. doi: 10.1042/BST0360791. Biochem Soc Trans. 2008. PMID: 18793138 Review. - Ubiquitin-dependent proteolysis and cell cycle control in yeast.
Chun KT, Mathias N, Goebl MG. Chun KT, et al. Prog Cell Cycle Res. 1996;2:115-27. doi: 10.1007/978-1-4615-5873-6_12. Prog Cell Cycle Res. 1996. PMID: 9552389 Review.
Cited by
- Rice ubiquitin-conjugating enzyme OsUbc13 negatively regulates immunity against pathogens by enhancing the activity of OsSnRK1a.
Liu J, Nie B, Yu B, Xu F, Zhang Q, Wang Y, Xu W. Liu J, et al. Plant Biotechnol J. 2023 Aug;21(8):1590-1610. doi: 10.1111/pbi.14059. Epub 2023 Apr 27. Plant Biotechnol J. 2023. PMID: 37102249 Free PMC article. - Systematic characterization of Brassica napus UBC13 genes involved in DNA-damage response and K63-linked polyubiquitination.
Kumasaruge I, Wen R, Wang L, Gao P, Peng G, Xiao W. Kumasaruge I, et al. BMC Plant Biol. 2023 Jan 12;23(1):24. doi: 10.1186/s12870-023-04035-y. BMC Plant Biol. 2023. PMID: 36631796 Free PMC article. - Regulation of Cdc42 protein turnover modulates the filamentous growth MAPK pathway.
González B, Cullen PJ. González B, et al. J Cell Biol. 2022 Dec 5;221(12):e202112100. doi: 10.1083/jcb.202112100. Epub 2022 Nov 9. J Cell Biol. 2022. PMID: 36350310 Free PMC article. - ALCAP2 inhibits lung adenocarcinoma cell proliferation, migration and invasion via the ubiquitination of β-catenin by upregulating the E3 ligase NEDD4L.
Zhang W, Zhang R, Zeng Y, Li Y, Chen Y, Zhou J, Zhang Y, Wang A, Zhu J, Liu Z, Yan Z, Huang JA. Zhang W, et al. Cell Death Dis. 2021 Jul 31;12(8):755. doi: 10.1038/s41419-021-04043-6. Cell Death Dis. 2021. PMID: 34330894 Free PMC article. - A selective transmembrane recognition mechanism by a membrane-anchored ubiquitin ligase adaptor.
Arines FM, Hamlin AJ, Yang X, Liu YJ, Li M. Arines FM, et al. J Cell Biol. 2021 Jan 4;220(1):e202001116. doi: 10.1083/jcb.202001116. J Cell Biol. 2021. PMID: 33351099 Free PMC article.
References
- Adams AEM, Botstein D, Drubin DG. A yeast actin-binding protein is encoded by SAC6, a gene found by suppression of an actin mutation. Science. 1989;243:231–233. - PubMed
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
- Attardi G, Schatz G. Biogenesis of mitochondria. Annu Rev Cell Biol. 1988;4:289–333. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases