Cloning and characterization of blaVIM, a new integron-borne metallo-beta-lactamase gene from a Pseudomonas aeruginosa clinical isolate - PubMed (original) (raw)
Cloning and characterization of blaVIM, a new integron-borne metallo-beta-lactamase gene from a Pseudomonas aeruginosa clinical isolate
L Lauretti et al. Antimicrob Agents Chemother. 1999 Jul.
Abstract
Production of a metallo-beta-lactamase activity was detected in a carbapenem-resistant Pseudomonas aeruginosa clinical isolate (isolate VR-143/97) from an Italian inpatient at the Verona University Hospital (northern Italy). The metallo-beta-lactamase determinant was isolated from a genomic library of VR-143/97, constructed in an Escherichia coli plasmid vector, by screening for clones with reduced susceptibility to imipenem. Sequencing of the cloned gene revealed that it encoded a new class B beta-lactamase that was named VIM-1. At the sequence level VIM-1 was rather divergent from the other class B enzymes (16.4 to 38.7% identity), overall being more similar to members of subclass B1 including the beta-lactamase II of Bacillus cereus (Bc-II), the Bacteroides fragilis CcrA, the Chryseobacterium meningosepticum BlaB, and the cassette-encoded IMP-1 enzymes. Among these, VIM-1 showed the highest degree of similarity to Bc-II. Similarly to blaIMP, blaVIM was also found to be carried on a gene cassette inserted into a class 1 integron. The blaVIM-containing integron was located on the chromosome of P. aeruginosa VR-143/97, and the metallo-beta-lactamase-encoding determinant was not transferable to E. coli by conjugation. Expression of the integron-borne blaVIM gene in E. coli resulted in a significant decrease in susceptibility to a broad array of beta-lactams (ampicillin, carbenicillin, piperacillin, mezlocillin, cefotaxime, cefoxitin, ceftazidime, cefoperazone, cefepime, and carbapenems), revealing a very broad substrate specificity of the VIM-1 enzyme.
Figures
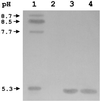
FIG. 1
Results of IEF analysis of crude extracts of P. aeruginosa VR-143/97 (lane 1), E. coli DH5α(pACYC184) (lane 2), E. coli DH5α(pAC2AL) (lane 3), and E. coli DH5α(pAC2IL) (lane 4). Approximately 10 μg of total protein was loaded in each lane.
FIG. 2
Physical map of the insert of plasmid pAC2AL and subcloning strategy. Thick lines represent cloned DNA, and thin lines correspond to vector sequences. Plasmids whose names begins with pAC are pACYC184 derivatives, while those whose name begins with pBC are pBC-SK derivatives. Production of metallo-β-lactamase activity (β-lact.) was assayed with crude extracts as described in Materials and Methods. B, _Bam_HI; C, _Cla_I; H, _Hin_dIII; Sa, _Sal_I; Sm, _Sma_I; S/B, _Sau_3AI/_Bam_HI junction; V, _Eco_RV; X, _Xho_I.
FIG. 3
Nucleotide sequence of the _bla_VIM gene cassette and flanking regions. Initiation codons of the various ORFs are indicated, and protein translation is reported below the sequence. The _bla_VIM cassette boundaries are indicated by vertical arrows. The conserved recombination core sites located at the cassette boundaries and the inverse core site are boxed. The internal 2L and 2R core sites of the 59-base element (36) are overlined with arrows.
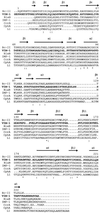
FIG. 4
Comparison of the VIM-1 sequence (VIM-1; boldfaced) with those of other molecular class B β-lactamases. Bc-II, β-lactamase II of B. cereus 569/H (13); BlaB, BlaB enzyme of C. meningosepticum CCUG4310 (32); IMP-1, IMP-1 enzyme encoded by the _bla_IMP gene cassette found in various gram-negative bacteria (2, 18, 25, 41); CcrA, CcrA enzyme of B. fragilis TAL3636 (30); CphA, CphA enzyme of A. hydrophila AE036 (21); L1, L1 enzyme of S. maltophilia IID1275 (38). Identical residues are indicated by an asterisk. Conserved amino acid substitutions are indicated by a colon. Secondary structure elements of Bc-II (5) are also indicated above the sequences. The numbering scheme refers to the Bc-II enzyme.
Similar articles
- Characterization of VIM-2, a carbapenem-hydrolyzing metallo-beta-lactamase and its plasmid- and integron-borne gene from a Pseudomonas aeruginosa clinical isolate in France.
Poirel L, Naas T, Nicolas D, Collet L, Bellais S, Cavallo JD, Nordmann P. Poirel L, et al. Antimicrob Agents Chemother. 2000 Apr;44(4):891-7. doi: 10.1128/AAC.44.4.891-897.2000. Antimicrob Agents Chemother. 2000. PMID: 10722487 Free PMC article. - Characterization of the metallo-beta-lactamase determinant of Acinetobacter baumannii AC-54/97 reveals the existence of bla(IMP) allelic variants carried by gene cassettes of different phylogeny.
Riccio ML, Franceschini N, Boschi L, Caravelli B, Cornaglia G, Fontana R, Amicosante G, Rossolini GM. Riccio ML, et al. Antimicrob Agents Chemother. 2000 May;44(5):1229-35. doi: 10.1128/AAC.44.5.1229-1235.2000. Antimicrob Agents Chemother. 2000. PMID: 10770756 Free PMC article. - Characterization of the new metallo-beta-lactamase VIM-13 and its integron-borne gene from a Pseudomonas aeruginosa clinical isolate in Spain.
Juan C, Beceiro A, Gutiérrez O, Albertí S, Garau M, Pérez JL, Bou G, Oliver A. Juan C, et al. Antimicrob Agents Chemother. 2008 Oct;52(10):3589-96. doi: 10.1128/AAC.00465-08. Epub 2008 Jul 21. Antimicrob Agents Chemother. 2008. PMID: 18644957 Free PMC article. - [Metallo-beta-lactamase producing bacteria].
Iyobe S. Iyobe S. Nihon Rinsho. 2001 Apr;59(4):701-6. Nihon Rinsho. 2001. PMID: 11304992 Review. Japanese. - Epidemiology and Characteristics of Metallo-β-Lactamase-Producing Pseudomonas aeruginosa.
Hong DJ, Bae IK, Jang IH, Jeong SH, Kang HK, Lee K. Hong DJ, et al. Infect Chemother. 2015 Jun;47(2):81-97. doi: 10.3947/ic.2015.47.2.81. Epub 2015 Jun 30. Infect Chemother. 2015. PMID: 26157586 Free PMC article. Review.
Cited by
- Epidemiology of resistance of carbapenemase-producing Klebsiella pneumoniae to ceftazidime-avibactam in a Chinese hospital.
Chen D, Xiao L, Hong D, Zhao Y, Hu X, Shi S, Chen F. Chen D, et al. J Appl Microbiol. 2022 Jan;132(1):237-243. doi: 10.1111/jam.15166. Epub 2021 Jul 22. J Appl Microbiol. 2022. PMID: 34053144 Free PMC article. - FIM-1, a new acquired metallo-β-lactamase from a Pseudomonas aeruginosa clinical isolate from Italy.
Pollini S, Maradei S, Pecile P, Olivo G, Luzzaro F, Docquier JD, Rossolini GM. Pollini S, et al. Antimicrob Agents Chemother. 2013 Jan;57(1):410-6. doi: 10.1128/AAC.01953-12. Epub 2012 Oct 31. Antimicrob Agents Chemother. 2013. PMID: 23114762 Free PMC article. - Bibliometric analysis of global scientific research on carbapenem resistance (1986-2015).
Sweileh WM, Shraim NY, Al-Jabi SW, Sawalha AF, AbuTaha AS, Zyoud SH. Sweileh WM, et al. Ann Clin Microbiol Antimicrob. 2016 Sep 23;15(1):56. doi: 10.1186/s12941-016-0169-6. Ann Clin Microbiol Antimicrob. 2016. PMID: 27663999 Free PMC article. - Purification and biochemical characterization of the VIM-1 metallo-beta-lactamase.
Franceschini N, Caravelli B, Docquier JD, Galleni M, Frère JM, Amicosante G, Rossolini GM. Franceschini N, et al. Antimicrob Agents Chemother. 2000 Nov;44(11):3003-7. doi: 10.1128/AAC.44.11.3003-3007.2000. Antimicrob Agents Chemother. 2000. PMID: 11036013 Free PMC article. - Genetic and biochemical characterization of FUS-1 (OXA-85), a narrow-spectrum class D beta-lactamase from Fusobacterium nucleatum subsp. polymorphum.
Voha C, Docquier JD, Rossolini GM, Fosse T. Voha C, et al. Antimicrob Agents Chemother. 2006 Aug;50(8):2673-9. doi: 10.1128/AAC.00058-06. Antimicrob Agents Chemother. 2006. PMID: 16870757 Free PMC article.
References
- Bush K. Metallo-β-lactamases: a class apart. Clin Infect Dis. 1998;27(Suppl. 1):S48–S53. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous