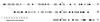Evidence that a plant virus switched hosts to infect a vertebrate and then recombined with a vertebrate-infecting virus - PubMed (original) (raw)
Evidence that a plant virus switched hosts to infect a vertebrate and then recombined with a vertebrate-infecting virus
M J Gibbs et al. Proc Natl Acad Sci U S A. 1999.
Abstract
There are several similarities between the small, circular, single-stranded-DNA genomes of circoviruses that infect vertebrates and the nanoviruses that infect plants. We analyzed circovirus and nanovirus replication initiator protein (Rep) sequences and confirmed that an N-terminal region in circovirus Reps is similar to an equivalent region in nanovirus Reps. However, we found that the remaining C-terminal region is related to an RNA-binding protein (protein 2C), encoded by picorna-like viruses, and we concluded that the sequence encoding this region of Rep was acquired from one of these single-stranded RNA viruses, probably a calicivirus, by recombination. This is clear evidence that a DNA virus has incorporated a gene from an RNA virus, and the fact that none of these viruses code for a reverse transcriptase suggests that another agent with this capacity was involved. Circoviruses were thought to be a sister-group of nanoviruses, but our phylogenetic analyses, which take account of the recombination, indicate that circoviruses evolved from a nanovirus. A nanovirus DNA was transferred from a plant to a vertebrate. This transferred DNA included the viral origin of replication; the sequence conservation clearly indicates that it maintained the ability to replicate. In view of these properties, we conclude that the transferred DNA was a kind of virus and the transfer was a host-switch. We speculate that this host-switch occurred when a vertebrate was exposed to sap from an infected plant. All characterized caliciviruses infect vertebrates, suggesting that the host-switch happened first and that the recombination took place in a vertebrate.
Figures
Figure 1
An alignment of Rep sequences from Banana bunchy top nanovirus Taiwanese isolate DNA1 (BBTV-T1), Coconut foliar decay nanovirus (CFDV), PCV type 1, and BFDV, together with the 2C-protein conserved region sequences from Norwalk calicivirus (NV) and the Feline calicivirus (FCV). The region in which the recombination probably occurred is marked with a solid-line box. Identities between the N-terminal sequences of circovirus and nanovirus Reps are marked with gray blocks, and those between calicivirus 2C-proteins and the C-terminal sequences of circovirus Reps are marked with black blocks. P-loop sequences are marked with dashed-line boxes.
Figure 2
Predicted secondary structures for geminivirus Reps (A), nanovirus Reps (B), and calicivirus 2C-proteins (C). Black arrows represent regions predicted to form β-strands, and gray helices represent regions predicted to form α-helices. Predictions were made from separate alignments of four geminivirus, ten nanovirus, and six calicivirus sequences. The position of the P-loop in each set of sequences is marked “P.” Dotted lines join structural elements that had matching positions when sequences were aligned.
Figure 3
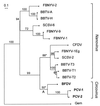
A maximum likelihood tree for the N-terminal region of the available nanovirus and circovirus Rep amino acid sequences (up to position 129, see Fig. 1). The equivalent sequences from seven geminiviruses were used as an out-group (marked “Gem”) to root the tree. Sequences: BBTV-A and -H, Banana bunchy top nanovirus isolates from Australia and Hawaii; BBTV-T1, -T2, and -T3, Taiwanese isolates DNAs 1, 2, and 3; BFDV, Psittacine beak and feather disease circovirus; CFDV, Coconut foliar decay nanovirus; FBNYV-1, -2, -9, and -1Eg, Faba bean necrotic yellows nanovirus components from isolates from Syria and Egypt; PCV-1 and -2, Porcine circovirus types 1 and 2; SCSV-2 and -6, Subterranean clover stunt nanovirus components 2 and 6. Bootstrap values are percentages from 10,000 neighbor-joining trees inferred from the amino acid sequences. Asterisks mark branches not found in the maximum likelihood or most parsimonious trees inferred from nucleotide sequences. Note that several nanovirus isolates have two or more distinct Rep genes carried by different genomic molecules.
Figure 4
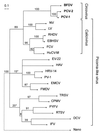
A maximum likelihood tree for the 2C-protein amino acid conserved sequences of picorna-like viruses and the equivalent region from the circovirus Reps (from position 178 to 313; see Fig. 1). The equivalent C-terminal regions of six nanovirus Reps were included in the analysis and these sequences are represented by the node marked “Nano.” The actual estimate for the length of the branch leading to the “Nano” node is double that shown. Sequences: BFDV, Psittacine beak and feather disease circovirus; CPMV, Cowpea mosaic comovirus, DCV, Drosophila virus C; EBHSV, European brown hare syndrome calicivirus; EMV, Encephalomyocarditis cardiovirus; EV-22, Echovirus 22; FCV, Feline calicivirus; FMDV, Foot and mouth disease aphthovirus; HAV, Hepatitis A hepatovirus; HRV-14, Human rhinovirus 14; HuCV-M, Human calicivirus Manchester isolate; IFV, Infectious flacherie virus; LV, Lordsdale calicivirus; NV, Norwalk calicivirus; PCV-1 and -2, Porcine circovirus types 1 and 2; PV-1, Poliovirus 1; PYFV, Parsnip yellow fleck sequivirus; RHDV, Rabbit hemorrhagic disease virus; RTSV, Rice tungro spherical waikavirus; TRSV, Tobacco ringspot nepovirus. Bootstrap values are percentages from 10,000 neighbor-joining trees inferred from the amino acid sequences. Asterisks mark branches not found in the maximum likelihood or most parsimonious trees inferred from nucleotide sequences.
Similar articles
- Sequence of porcine circovirus DNA: affinities with plant circoviruses.
Meehan BM, Creelan JL, McNulty MS, Todd D. Meehan BM, et al. J Gen Virol. 1997 Jan;78 ( Pt 1):221-7. doi: 10.1099/0022-1317-78-1-221. J Gen Virol. 1997. PMID: 9010307 - The master rep concept in nanovirus replication: identification of missing genome components and potential for natural genetic reassortment.
Timchenko T, Katul L, Sano Y, de Kouchkovsky F, Vetten HJ, Gronenborn B. Timchenko T, et al. Virology. 2000 Aug 15;274(1):189-95. doi: 10.1006/viro.2000.0439. Virology. 2000. PMID: 10936099 - Nanoviruses: genome organisation and protein function.
Gronenborn B. Gronenborn B. Vet Microbiol. 2004 Feb 4;98(2):103-9. doi: 10.1016/j.vetmic.2003.10.015. Vet Microbiol. 2004. PMID: 14741122 Review. - Negative-strand RNA viruses: the plant-infecting counterparts.
Kormelink R, Garcia ML, Goodin M, Sasaya T, Haenni AL. Kormelink R, et al. Virus Res. 2011 Dec;162(1-2):184-202. doi: 10.1016/j.virusres.2011.09.028. Epub 2011 Sep 22. Virus Res. 2011. PMID: 21963660 Review.
Cited by
- Evidence of unique genotypes of beak and feather disease virus in southern Africa.
Heath L, Martin DP, Warburton L, Perrin M, Horsfield W, Kingsley C, Rybicki EP, Williamson AL. Heath L, et al. J Virol. 2004 Sep;78(17):9277-84. doi: 10.1128/JVI.78.17.9277-9284.2004. J Virol. 2004. PMID: 15308722 Free PMC article. - Association and host selectivity in multi-host pathogens.
Malpica JM, Sacristán S, Fraile A, García-Arenal F. Malpica JM, et al. PLoS One. 2006 Dec 20;1(1):e41. doi: 10.1371/journal.pone.0000041. PLoS One. 2006. PMID: 17183670 Free PMC article. - An evolutionary analysis of the Secoviridae family of viruses.
Thompson JR, Kamath N, Perry KL. Thompson JR, et al. PLoS One. 2014 Sep 2;9(9):e106305. doi: 10.1371/journal.pone.0106305. eCollection 2014. PLoS One. 2014. PMID: 25180860 Free PMC article. - Development of real-time PCR assay for detection of porcine circovirus-like virus P1 in domestic pigs in China.
He KW, Wen LB, Wang YS, Lu CP. He KW, et al. BMC Vet Res. 2015 Sep 24;11:240. doi: 10.1186/s12917-015-0509-3. BMC Vet Res. 2015. PMID: 26404908 Free PMC article. - Evolutionary dynamics of genome segmentation in multipartite viruses.
Iranzo J, Manrubia SC. Iranzo J, et al. Proc Biol Sci. 2012 Sep 22;279(1743):3812-9. doi: 10.1098/rspb.2012.1086. Epub 2012 Jul 4. Proc Biol Sci. 2012. PMID: 22764164 Free PMC article.
References
- Morse S S. In: The Evolutionary Biology of Viruses. Morse S S, editor. New York: Raven; 1994. pp. 325–335.
- Sharp P M, Robertson D L, Hahn B H. Philos Trans R Soc London B. 1995;349:41–47. - PubMed
- Webster R G, Bean W J, Gorman O T. In: Molecular Basis of Virus Evolution. Gibbs A J, Calisher C H, Garcia-Arenal F, editors. Cambridge, U.K.: Cambridge Univ. Press; 1995. pp. 531–543.
- Gibbs A J, Keese P L, Gibbs M J, Garcia-Arenal F. In: Origin and Evolution of Viruses. Domingo E, Webster R, Holland J, editors. London: Academic; 1998. pp. 263–285.
- Murphy F A, Fauquet C M, Bishop D H L, Ghabrial S A, Jarvis A W, Martelli G P, Mayo M A, Summers M D. Virus Taxonomy: Classification and Nomenclature of Viruses. Vienna: Springer; 1995. p. 586.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous