Saccharomyces cerevisiae MPS2 encodes a membrane protein localized at the spindle pole body and the nuclear envelope - PubMed (original) (raw)
Saccharomyces cerevisiae MPS2 encodes a membrane protein localized at the spindle pole body and the nuclear envelope
M C Muñoz-Centeno et al. Mol Biol Cell. 1999 Jul.
Free PMC article
Abstract
The MPS2 (monopolar spindle two) gene is one of several genes required for the proper execution of spindle pole body (SPB) duplication in the budding yeast Saccharomyces cerevisiae (). We report here that the MPS2 gene encodes an essential 44-kDa protein with two putative coiled-coil regions and a hydrophobic sequence. Although MPS2 is required for normal mitotic growth, some null strains can survive; these survivors exhibit slow growth and abnormal ploidy. The MPS2 protein was tagged with nine copies of the myc epitope, and biochemical fractionation experiments show that it is an integral membrane protein. Visualization of a green fluorescent protein (GFP) Mps2p fusion protein in living cells and indirect immunofluorescence microscopy of 9xmyc-Mps2p revealed a perinuclear localization with one or two brighter foci of staining corresponding to the SPB. Additionally, immunoelectron microscopy shows that GFP-Mps2p localizes to the SPB. Our analysis suggests that Mps2p is required as a component of the SPB for insertion of the nascent SPB into the nuclear envelope.
Figures
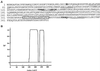
Figure 1
Features of the MPS2 encoded protein. (A) Deduced amino acid sequence of the MPS2 encoded protein. The putative transmembrane segment (residues 311–327) is boxed. Potential Cdc28p phosphorylation sites are indicated in bold letters, and potential destruction box sequences are underlined. The point mutation found in the mps2–1 allele at amino acid 39 is highlighted (E to K). (B) Coiled-coil probability plot. The _x_-axis is the position of Mps2p amino acids and the _y_-axis is the probability (p) of being in a coiled-coil using the COILS program (
http://www.ch.embnet.org/software/COILS
form.html), based on the Lupas algorithm (Lupas et al., 1991).
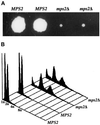
Figure 2
Deletion of MPS2 results in abnormal ploidy. (A) A representative tetrad from the mps2Δ(116–387)::HIS3/MPS2 strain (AM610) grown for 14 d at 23°C. (B) Histogram of relative DNA content from each of the spore clones shown in A.
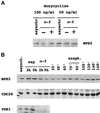
Figure 3
Mps2p levels show little fluctuation during the cell cycle. (A) An overnight culture of MCL 94 cells containing pTet-9xmycN-MPS2 grown in the presence of 5 μg/ml doxycycline (repressing conditions) was diluted in the same selective medium containing 50 or 100 ng/ml doxycycline to allow a low constitutive expression from the tet promoter. Cells were grown overnight to early log phase at 30°C (asynchr.) and then divided into two separate cultures: one culture was left untreated (−) and the other was arrested in the G1 phase with α-factor (0.2 μM final concentration) for 2 h (+). Equal quantities of total cell protein were then subjected to SDS-PAGE, and proteins were transferred to membranes to determine the levels of 9xmycN-Mps2p by immunoblot analysis. 9xmycN-Mps2p is absent when cells are grown in the presence of fully repressing concentrations of doxycycline (5 μg/ml) or from cells that do not contain the 9xmycN-MPS2 plasmid. The G1 arrest was verified by flow cytometric analysis. (B) Wild-type cells expressing both 9xmycN-Mps2p and Pds1-HAp (MCL123) were grown to early log phase at 24°C (asynchr.) and then divided into two separate cultures: one culture was left untreated (exp) and the other was arrested in the G1 phase with α-factor (0.2 μM) for 2 or 5 h. After 2 h, >90% of the cells were already arrested in G1 phase as determined by flow cytometry analysis, as were cells treated with α-factor for 5 h. After this time, cells were released from the arrest by washing out the α-factor. Samples were taken at the indicated time points and analyzed for 9xmycN-Mps2p, Cdc28p, and Pds1-HAp by immunoblot analysis. Cell and nuclear morphology and DNA content were analyzed by propidium iodide staining.
Figure 4
9xmycN-Mps2p fractionates as an integral membrane protein. A whole-cell extract was prepared from cells expressing 9xmycN-Mps2p from its normal promoter (MCL120) by gentle spheroplast lysis (see MATERIALS AND METHODS) and then fractionated as described in MATERIALS AND METHODS. (A) Cell fractionation: the same amount of protein from the whole-cell extract and S13, P13, P100, and S100 fractions were subjected to SDS-PAGE and immunoblotted with anti-myc, anti-Sec61p, anti-Dpm1p, and anti-hexokinase antibodies successively. (B) Solubilization of Mps2p from the P13 fraction prepared from MCL120. The pellet was extracted with lysis buffer (control) or lysis buffer containing 1 M NaCl, 50 mM Tris (pH 7.5)-10 mM EDTA, 0.2 M Na2CO3 (pH 11), 1% Triton X-100, or 6 M urea as described in MATERIALS AND METHODS. The mixtures were incubated on ice for 10 min and then centrifuged to produce a pellet fraction (P) and a supernatant fraction (S). Equivalent samples of the pellet and supernatant were analyzed by immunoblotting with anti-myc or anti-Sec61 antibodies. (C) Total solubilization of Mps2p protein from the P13. The P13 pellet was extracted with lysis buffer (control) or lysis buffer containing 1% Triton X-100, 1% Triton X-100 plus 6 M urea, or 1% Triton X-100 plus 1 M NaCl. Pellet (P) and supernatant (S) were separated and analyzed by immunoblotting.
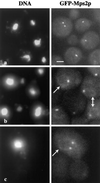
Figure 5
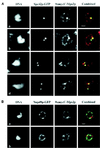
Localization of GFP-Mps2p in living cells. Several cells are shown in a, in which GFP-Mps2p (strain MCL175) localizes in one or two discrete foci coincident with the nuclear DNA. The cell with two distinct spots of fluorescence is a small budded cell. The cells shown in b reveal that GFP-Mps2p also localizes to the nuclear periphery. Several cells are shown with one bright spot and a fainter signal at the nuclear periphery (arrows). A higher magnification of a budded cell is shown in c with two bright spots and a perinuclear localization. The GFP-Mps2p was visualized via autofluorescence of GFP, and the DNA was visualized with Hoechst. A single plane of focus is shown. Bar, 1 μm.
Figure 6
9xmycC-Mps2p colocalizes with Spc42p-GFP, but not Nup49p-GFP. (A) A strain expressing both 9xmycC-Mps2p and Spc42p-GFP (SMY28–2b) was prepared for indirect immunofluorescence microscopy (see MATERIALS AND METHODS). The Spc42p-GFP was visualized via autofluorescence of GFP, the 9xmycC-Mps2p was detected using a Texas Red-conjugated 2° antibody, and the DNA was visualized with DAPI. Four different cells are shown at a high magnification to show the detail of colocalization. The bright spots of 9xmycC-Mps2p (red) and the Spc42p-GFP (green) signals overlap, as seen in the combined image (yellow). The Spc42-GFP and 9xmycC-Mps2p signals at the lower SPBs in images b and d do not completely overlap in these focal planes; however, Spc42p and Mps2p are not expected to be in the same exact position in the organelle. In addition, the amount of overlap depends on the ability to indirectly label Mps2p at any given SPB. (B) Two different cells from a strain expressing both 9xmycC-Mps2p and Nup49p-GFP (SMY1892) are shown at a high magnification. There are bright spots of 9xmycC-Mps2p fluorescence observed along the nuclear periphery that are devoid of Nup49p-GFP signal. All panels in this Figure represent a single optical plane that was deconvolved using the Slidebook software package (see MATERIALS AND METHODS). Bar, 1 μm in both panels.
Figure 7
GFP-Mps2p localizes to the SPB. Cells expressing GFP-Mps2p were prepared for immunoelectron microscopy using anti-GFP antibodies and colloidal gold-conjugated secondary antibodies. Thin sections of four different cells are shown. C, cytoplasm; N, nucleoplasm; NE, nuclear envelope; MTs, microtubules; SPB, spindle pole body. Arrowheads show the localization of individual gold particles in b and d. Gold particles were observed in all 12 SPBs examined. Bar, 0.1 μm.
Similar articles
- The Saccharomyces cerevisiae spindle pole body (SPB) component Nbp1p is required for SPB membrane insertion and interacts with the integral membrane proteins Ndc1p and Mps2p.
Araki Y, Lau CK, Maekawa H, Jaspersen SL, Giddings TH Jr, Schiebel E, Winey M. Araki Y, et al. Mol Biol Cell. 2006 Apr;17(4):1959-70. doi: 10.1091/mbc.e05-07-0668. Epub 2006 Jan 25. Mol Biol Cell. 2006. PMID: 16436507 Free PMC article. - The Sad1-UNC-84 homology domain in Mps3 interacts with Mps2 to connect the spindle pole body with the nuclear envelope.
Jaspersen SL, Martin AE, Glazko G, Giddings TH Jr, Morgan G, Mushegian A, Winey M. Jaspersen SL, et al. J Cell Biol. 2006 Aug 28;174(5):665-75. doi: 10.1083/jcb.200601062. Epub 2006 Aug 21. J Cell Biol. 2006. PMID: 16923827 Free PMC article. - A ternary membrane protein complex anchors the spindle pole body in the nuclear envelope in budding yeast.
Kupke T, Malsam J, Schiebel E. Kupke T, et al. J Biol Chem. 2017 May 19;292(20):8447-8458. doi: 10.1074/jbc.M117.780601. Epub 2017 Mar 29. J Biol Chem. 2017. PMID: 28356353 Free PMC article. - Composition of the spindle pole body of Saccharomyces cerevisiae and the proteins involved in its duplication.
Helfant AH. Helfant AH. Curr Genet. 2002 Feb;40(5):291-310. doi: 10.1007/s00294-001-0263-x. Epub 2001 Dec 8. Curr Genet. 2002. PMID: 11935220 Review. - Nuclear envelope insertion of spindle pole bodies and nuclear pore complexes.
Jaspersen SL, Ghosh S. Jaspersen SL, et al. Nucleus. 2012 May-Jun;3(3):226-36. doi: 10.4161/nucl.20148. Epub 2012 May 1. Nucleus. 2012. PMID: 22572959 Free PMC article. Review.
Cited by
- LINC complexes in health and disease.
Méjat A, Misteli T. Méjat A, et al. Nucleus. 2010 Jan-Feb;1(1):40-52. doi: 10.4161/nucl.1.1.10530. Nucleus. 2010. PMID: 21327104 Free PMC article. Review. - Mitotic spindle form and function.
Winey M, Bloom K. Winey M, et al. Genetics. 2012 Apr;190(4):1197-224. doi: 10.1534/genetics.111.128710. Genetics. 2012. PMID: 22491889 Free PMC article. - Mps2 links Csm4 and Mps3 to form a telomere-associated LINC complex in budding yeast.
Fan J, Jin H, Koch BA, Yu HG. Fan J, et al. Life Sci Alliance. 2020 Sep 23;3(12):e202000824. doi: 10.26508/lsa.202000824. Print 2020 Dec. Life Sci Alliance. 2020. PMID: 32967926 Free PMC article. - Sec66-Dependent Regulation of Yeast Spindle-Pole Body Duplication Through Pom152.
Katta SS, Chen J, Gardner JM, Friederichs JM, Smith SE, Gogol M, Unruh JR, Slaughter BD, Jaspersen SL. Katta SS, et al. Genetics. 2015 Dec;201(4):1479-95. doi: 10.1534/genetics.115.178012. Epub 2015 Oct 28. Genetics. 2015. PMID: 26510791 Free PMC article. - Extranuclear Structural Components that Mediate Dynamic Chromosome Movements in Yeast Meiosis.
Lee CY, Bisig CG, Conrad MM, Ditamo Y, Previato de Almeida L, Dresser ME, Pezza RJ. Lee CY, et al. Curr Biol. 2020 Apr 6;30(7):1207-1216.e4. doi: 10.1016/j.cub.2020.01.054. Epub 2020 Feb 13. Curr Biol. 2020. PMID: 32059771 Free PMC article.
References
- Amon A, Irniger S, Nasmyth K. Closing the cell cycle circle in yeast: G2 cyclin proteolysis initiated at mitosis persists until the activation of G1 cyclins in the next cycle. Cell. 1994;77:1037–1050. - PubMed
- Boeke JD, Trueheart J, Natsoulis G, Fink GR. 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 1987;154:164–175. - PubMed
- Brachmann CB, Davies A, Cost GJ, Caputo E, Li J, Hieter P, Boeke JD. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–132. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases