The furin protease cleavage recognition sequence of Sindbis virus PE2 can mediate virion attachment to cell surface heparan sulfate - PubMed (original) (raw)
The furin protease cleavage recognition sequence of Sindbis virus PE2 can mediate virion attachment to cell surface heparan sulfate
W B Klimstra et al. J Virol. 1999 Aug.
Abstract
Cell culture-adapted Sindbis virus strains attach to heparan sulfate (HS) receptors during infection of cultured cells (W. B. Klimstra, K. D. Ryman, and R. E. Johnston, J. Virol. 72:7357-7366, 1998). At least three E2 glycoprotein mutations (E2 Arg 1, E2 Lys 70, and E2 Arg 114) can independently confer HS attachment in the background of the consensus sequence Sindbis virus (TR339). In the studies reported here, we have investigated the mechanism by which the E2 Arg 1 mutation confers HS-dependent binding. Substitution of Arg for Ser at E2 1 resulted in a significant reduction in the efficiency of PE2 cleavage, yielding virus particles containing a mixture of PE2 and mature E2. Presence of PE2 was associated with an increase in HS-dependent attachment to cells and efficient attachment to heparin-agarose beads, presumably because the furin recognition site for PE2 cleavage also represents a candidate HS binding sequence. A comparison of mutants with partially or completely inhibited PE2 cleavage demonstrated that efficiency of cell binding was correlated with the amount of PE2 in virus particles. Viruses rendered cleavage defective due to deletions of portions or all of the furin cleavage sequence attached very poorly to cells, indicating that an intact furin cleavage sequence was specifically required for PE2-mediated attachment to cells. In contrast, a virus containing a partial deletion was capable of efficient binding to heparin-agarose beads, suggesting different requirements for heparin bead and cell surface HS binding. Furthermore, virus produced in C6/36 mosquito cells, which cleave PE2 more efficiently than BHK cells, exhibited a reduction in cell attachment efficiency correlated with reduced content of PE2 in particles. Taken together, these results strongly argue that the XBXBBX (B, basic; X, hydrophobic) furin protease recognition sequence of PE2 can mediate the binding of PE2-containing Sindbis viruses to HS. This sequence is very similar to an XBBXBX heparin-HS interaction consensus sequence. The attachment of furin protease cleavage sequences to HS may have relevance to other viruses whose attachment proteins are cleaved during maturation at positively charged recognition sequences.
Figures
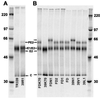
FIG. 1
SDS-PAGE analysis of purified radiolabeled virus particles; 5.0 × 104 cpm of virus was loaded in each lane. Molecular mass markers (kDa) are shown in the extreme right-hand and left-hand lanes.
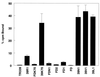
FIG. 2
Binding of radiolabeled purified viruses to CHO cells. The bars represent averages of triplicate binding assays with 105 cpm of virus and ∼106 cells per reaction. Each set of triplicates was repeated at least twice. The error bars represent standard deviations.
FIG. 3
Binding of viruses to CHO cells without (solid bars) or with (hatched bars) previous digestion with heparinase I. The cells were either digested with 8 U of heparinase I (in phosphate-buffered saline with 0.1% BSA/ml) or mock-digested (phosphate-buffered saline with 0.1% BSA only) for 1 h at 37°C followed by processing for attachment assays as described in Materials and Methods. The bars represent averages of triplicate binding assays with 105 cpm of virus and ∼106 cells per reaction. Each set of triplicates was repeated at least twice. The error bars represent standard deviations.
FIG. 4
Binding of radiolabeled viruses prepared in BHK cells or C6/36 cells to CHO cells. The bars represent averages of triplicate binding assays with 105 cpm of virus and ∼106 cells per reaction. Each set of triplicates was repeated at least twice. The error bars represent standard deviations.
Similar articles
- Adaptation of Sindbis virus to BHK cells selects for use of heparan sulfate as an attachment receptor.
Klimstra WB, Ryman KD, Johnston RE. Klimstra WB, et al. J Virol. 1998 Sep;72(9):7357-66. doi: 10.1128/JVI.72.9.7357-7366.1998. J Virol. 1998. PMID: 9696832 Free PMC article. - Differential processing of sindbis virus glycoprotein PE2 in cultured vertebrate and arthropod cells.
Heidner HW, Knott TA, Johnston RE. Heidner HW, et al. J Virol. 1996 Mar;70(3):2069-73. doi: 10.1128/JVI.70.3.2069-2073.1996. J Virol. 1996. PMID: 8627739 Free PMC article. - Attenuation of Sindbis virus variants incorporating uncleaved PE2 glycoprotein is correlated with attachment to cell-surface heparan sulfate.
Ryman KD, Klimstra WB, Johnston RE. Ryman KD, et al. Virology. 2004 Apr 25;322(1):1-12. doi: 10.1016/j.virol.2004.01.003. Virology. 2004. PMID: 15063111 - Maturation of HIV envelope glycoprotein precursors by cellular endoproteases.
Moulard M, Decroly E. Moulard M, et al. Biochim Biophys Acta. 2000 Nov 10;1469(3):121-32. doi: 10.1016/s0304-4157(00)00014-9. Biochim Biophys Acta. 2000. PMID: 11063880 Review. - The SARS-CoV-2 Entry Inhibition Mechanisms of Serine Protease Inhibitors, OM-85, Heparin and Soluble HS Might Be Linked to HS Attachment Sites.
Cheudjeu A. Cheudjeu A. Molecules. 2022 Mar 17;27(6):1947. doi: 10.3390/molecules27061947. Molecules. 2022. PMID: 35335311 Free PMC article. Review.
Cited by
- Dancing with the Devil: A Review of the Importance of Host RNA-Binding Proteins to Alphaviral RNAs during Infection.
Westcott CE, Isom CM, Karki D, Sokoloski KJ. Westcott CE, et al. Viruses. 2023 Jan 5;15(1):164. doi: 10.3390/v15010164. Viruses. 2023. PMID: 36680204 Free PMC article. Review. - Redirecting lentiviral vectors pseudotyped with Sindbis virus-derived envelope proteins to DC-SIGN by modification of N-linked glycans of envelope proteins.
Morizono K, Ku A, Xie Y, Harui A, Kung SK, Roth MD, Lee B, Chen IS. Morizono K, et al. J Virol. 2010 Jul;84(14):6923-34. doi: 10.1128/JVI.00435-10. Epub 2010 May 19. J Virol. 2010. PMID: 20484510 Free PMC article. - Electrostatics Drive the Molecular Chaperone BiP to Preferentially Bind Oligomerized States of a Client Protein.
Deans EE, Kotler JLM, Wei WS, Street TO. Deans EE, et al. J Mol Biol. 2022 Jul 15;434(13):167638. doi: 10.1016/j.jmb.2022.167638. Epub 2022 May 18. J Mol Biol. 2022. PMID: 35597552 Free PMC article. - Resuscitating mutations in a furin cleavage-deficient mutant of the flavivirus tick-borne encephalitis virus.
Elshuber S, Mandl CW. Elshuber S, et al. J Virol. 2005 Sep;79(18):11813-23. doi: 10.1128/JVI.79.18.11813-11823.2005. J Virol. 2005. PMID: 16140758 Free PMC article. - Requirement of a functional ion channel for Sindbis virus glycoprotein transport, CPV-II formation, and efficient virus budding.
Elmasri Z, Negi V, Kuhn RJ, Jose J. Elmasri Z, et al. PLoS Pathog. 2022 Oct 3;18(10):e1010892. doi: 10.1371/journal.ppat.1010892. eCollection 2022 Oct. PLoS Pathog. 2022. PMID: 36191050 Free PMC article.
References
- Barbouche R, Sabatier J M, Fenouillet E. An anti-HIV peptide construct derived from the cleavage region of the Env precursor acts on Env fusogenicity through the presence of a functional cleavage sequence. Virology. 1998;247:137–143. - PubMed
- Cardin A D, Weintraub H J R. Molecular modeling of protein-glycosaminoglycan interactions. Arteriosclerosis. 1989;9:21–32. - PubMed
- Chen Y, Maguire T, Hileman R E, Fromm J R, Esko J D, Linhardt R J, Marks R M. Dengue virus infectivity depends on envelope protein binding to target cell heparan sulfate. Nat Med. 1997;3:866–871. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- F32-AI09015/AI/NIAID NIH HHS/United States
- T32 AI007151/AI/NIAID NIH HHS/United States
- T32 AI007419/AI/NIAID NIH HHS/United States
- AI22186/AI/NIAID NIH HHS/United States
- T32 AI07419/AI/NIAID NIH HHS/United States
- R01 AI022186/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources