Comparison of molecular methods for typing Vibrio parahaemolyticus - PubMed (original) (raw)
Comparative Study
Comparison of molecular methods for typing Vibrio parahaemolyticus
S Marshall et al. J Clin Microbiol. 1999 Aug.
Abstract
An outbreak of Vibrio parahaemolyticus gastroenteritis on Canada's west coast in 1997 emphasized the need to develop molecular methods for differentiation and typing of these organisms. Isolates were analyzed by enterobacterial repetitive intergenic consensus sequence (ERIC) PCR, detection of restriction fragment length polymorphisms (RFLP) in rRNA genes (ribotyping), pulsed-field gel electrophoresis (PFGE), and RFLP analysis of the genetic locus encoding the polar flagellum (Fla locus RFLP analysis). ERIC PCR and ribotyping were the most informative typing methods, especially when used together, while Fla locus RFLP analysis was the least discriminatory. PFGE exhibited good discrimination but suffered from a high incidence of DNA degradation. ERIC PCR and ribotyping will be useful for the evaluation of genetic and epidemiological relationships among V. parahaemolyticus strains.
Figures
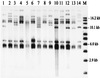
FIG. 1
Representative ribotype patterns of _Bgl_I-digested DNA. Pattern designations are given in parentheses. Lane 1, T97-31 (A); lane 2, T97-32 (B); lane 3, T97-33 (C); lane 4, T97-54 (D); lane 5, T97-57 (degraded DNA from isolate exhibiting pattern E in subsequent analysis); lane 6, T97-45 (F); lane 7, T97-58 (G); lane 8, T97-60 (H); lane 9, T97-52 (I); lane 10, T97-61 (J); lane 11, T97-65 (K); lane 12, T97-66 (L); lane 13, T97-472 (M); lane 14, T97-474 (N); lane M, _Pvu_II-digested supercoiled DNA, with sizes of selected standard bands shown at the right.
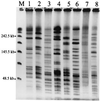
FIG. 2
Representative PFGE patterns for V. parahaemolyticus chromosomal DNA digested with _Apa_I. Pattern designations are given in parentheses. Lane M, λ DNA concatemer size standard, with sizes of selected bands given at the left; lane 1, T97-31 (A1); lane 2, T97-34 (A2); lane 3, T97-450 (B); lane 4, T97-42 (C); lane 5, T97-45 (D); lane 6, T97-52 (E); lane 7, T97-61 (I); lane 8, T97-65 (K).
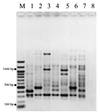
FIG. 3
ERIC 1R PCR profiles from selected V. parahaemolyticus isolates. Pattern designations are in parentheses. Lane M, 100-bp ladder; lane 1, T97-31 (A1); lane 2, T97-32 (B1); lane 3, T97-33 (C1); lane 4, T97-56 (D); lane 5, T97-65 (G); lane 6, T97-450 (I); lane 7, T97-474 (L); lane 8, negative control.
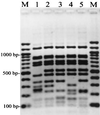
FIG. 4
RFLP patterns obtained by _Rsa_I digestion of the product of PCR of the V. parahaemolyticus Fla locus. Pattern designations are given in parentheses. Lanes M, 100-bp ladder; lane 1, T97-31 (A); lane 2, T97-32 (B); lane 3, T97-33 (C); lane 4, T97-58 (D); lane 5, T97-56 (E).
Similar articles
- Molecular typing of Vibrio parahaemolyticus isolates from the middle-east coastline of China.
Chen W, Xie Y, Xu J, Wang Q, Gu M, Yang J, Zhou M, Wang D, Shi C, Shi X. Chen W, et al. Int J Food Microbiol. 2012 Feb 15;153(3):402-12. doi: 10.1016/j.ijfoodmicro.2011.12.001. Epub 2011 Dec 19. Int J Food Microbiol. 2012. PMID: 22225982 - Evaluation of typing of vibrio parahaemolyticus by three PCR methods using specific primers.
Wong HC, Lin CH. Wong HC, et al. J Clin Microbiol. 2001 Dec;39(12):4233-40. doi: 10.1128/JCM.39.12.4233-4240.2001. J Clin Microbiol. 2001. PMID: 11724826 Free PMC article. - Molecular typing of Vibrio parahaemolyticus isolated from seafood harvested along the south-west coast of India.
Bhowmick PP, Khushiramani R, Raghunath P, Karunasagar I, Karunasagar I. Bhowmick PP, et al. Lett Appl Microbiol. 2008 Feb;46(2):198-204. doi: 10.1111/j.1472-765X.2007.02304.x. Epub 2007 Dec 20. Lett Appl Microbiol. 2008. PMID: 18179449 - [Homology analysis of Vibrio parahaemolyticus isolated by MPN method using pulsed-field gel electrophoresis and automated ribotyping].
Li W, Wang X, Pei X, Zhu H, Guo Y. Li W, et al. Wei Sheng Yan Jiu. 2010 May;39(3):374-80. Wei Sheng Yan Jiu. 2010. PMID: 20568474 Chinese.
Cited by
- Molecular fingerprinting of Clostridium difficile isolates: pulsed-field gel electrophoresis versus amplified fragment length polymorphism.
Klaassen CH, van Haren HA, Horrevorts AM. Klaassen CH, et al. J Clin Microbiol. 2002 Jan;40(1):101-4. doi: 10.1128/JCM.40.1.101-104.2002. J Clin Microbiol. 2002. PMID: 11773100 Free PMC article. - Phenotypic and genotypic characterization of Canadian clinical isolates of Vibrio parahaemolyticus collected from 2000 to 2009.
Banerjee SK, Kearney AK, Nadon CA, Peterson CL, Tyler K, Bakouche L, Clark CG, Hoang L, Gilmour MW, Farber JM. Banerjee SK, et al. J Clin Microbiol. 2014 Apr;52(4):1081-8. doi: 10.1128/JCM.03047-13. Epub 2014 Jan 22. J Clin Microbiol. 2014. PMID: 24452166 Free PMC article. - Population structure of clinical Vibrio parahaemolyticus from 17 coastal countries, determined through multilocus sequence analysis.
Han D, Tang H, Lu J, Wang G, Zhou L, Min L, Han C. Han D, et al. PLoS One. 2014 Sep 16;9(9):e107371. doi: 10.1371/journal.pone.0107371. eCollection 2014. PLoS One. 2014. PMID: 25225911 Free PMC article. - Pathogenic characteristics of the Vibrio parahaemolyticus which caused a gastroenteritis outbreak event in Huzhou.
Wu X, Zhu Y, Yan W, Zhang P, Chen L. Wu X, et al. FEMS Microbiol Lett. 2024 Jan 9;371:fnad130. doi: 10.1093/femsle/fnad130. FEMS Microbiol Lett. 2024. PMID: 38066691 Free PMC article. - Multiple-locus variable-number tandem-repeat analysis for clonal identification of Vibrio parahaemolyticus isolates by using capillary electrophoresis.
Harth-Chu E, Espejo RT, Christen R, Guzmán CA, Höfle MG. Harth-Chu E, et al. Appl Environ Microbiol. 2009 Jun;75(12):4079-88. doi: 10.1128/AEM.02729-08. Epub 2009 Apr 17. Appl Environ Microbiol. 2009. PMID: 19376898 Free PMC article.
References
- Blake P A, Weaver R E, Hollis D G. Diseases of humans (other than cholera) caused by vibrios. Annu Rev Microbiol. 1980;34:341–367. - PubMed
- Dalsgaard A, Serichantalergs O, Echeverria P. Restriction fragment length polymorphisms (RFLP) of cholera toxin genes in Vibrio cholerae O139 recovered from patients in Thailand, India and Bangladesh. Scand J Infect Dis. 1995;27:585–588. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources