PER and TIM inhibit the DNA binding activity of a Drosophila CLOCK-CYC/dBMAL1 heterodimer without disrupting formation of the heterodimer: a basis for circadian transcription - PubMed (original) (raw)
PER and TIM inhibit the DNA binding activity of a Drosophila CLOCK-CYC/dBMAL1 heterodimer without disrupting formation of the heterodimer: a basis for circadian transcription
C Lee et al. Mol Cell Biol. 1999 Aug.
Abstract
The Drosophila CLOCK (dCLOCK) and CYCLE (CYC) (also referred to as dBMAL1) proteins are members of the basic helix-loop-helix PAS (PER-ARNT-SIM) superfamily of transcription factors and are required for high-level expression of the circadian clock genes period (per) and timeless (tim). Several lines of evidence indicate that PER, TIM, or a PER-TIM heterodimer somehow inhibit the transcriptional activity of a putative dCLOCK-CYC complex, generating a negative-feedback loop that is a core element of the Drosophila circadian oscillator. In this report we show that PER and/or TIM inhibits the binding of a dCLOCK-CYC heterodimer to an E-box-containing DNA fragment that is present in the 5' nontranscribed region of per and acts as a circadian enhancer element. Surprisingly, inhibition of this DNA binding activity by PER, TIM, or both is not accompanied by disruption of the association between dCLOCK and CYC. The results suggest that the interaction of PER, TIM, or both with the dCLOCK-CYC heterodimer induces a conformational change or masks protein regions in the heterodimer, leading to a reduction in DNA binding activity. Together with other findings, our results strongly suggest that daily cycles in the association of PER and TIM with the dCLOCK-CYC complex probably contribute to rhythmic expression of per and tim.
Figures
FIG. 1
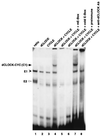
dCLOCK/CYC-mediated binding to an E box present in a per circadian regulatory element. Reticulocyte lysates containing 10 fmol each of dCLOCK (lane 2), CYC (lane 3), or a mixture of the two (lanes 4 to 8) were incubated with either the wild-type pwtE oligonucleotide (lanes 1 to 4 and 6 to 8) or the pmE E-box mutant version (lane 5). Subsequently, mixtures were resolved by electrophoresis on native polyacrylamide gels followed by autoradiography. The dCLOCK/CYC specific mobility shift (C1) is indicated by the arrow (left). Also indicated are two complexes, termed E1 (solid arrowhead) and E2 (open arrowhead), that are observed when using nonprogrammed reticulocyte lysate (lane 1). Addition of a 100-fold molar excess (1 μg) of cold wild-type E-box DNA abolished detection of C1, E1, and E2 (lane 6), whereas addition of antibodies (Ab) to dCLOCK specifically inhibited C1 (lanes 7 and 8). The “free” unbound radiolabeled DNA probe is at the bottom of the gel.
FIG. 2
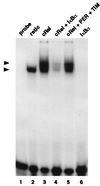
PER and TIM inhibit the DNA binding activity of dCLOCK/CYC. (A) Reticulocyte lysates containing dCLOCK (10 fmol), CYC (10 fmol), PER (20 fmol), and TIM (20 fmol) in different combinations (indicated at the top) were incubated with the pwtE [32P]DNA fragment and complexes visualized as described in the legend to figure 1. E2 is inhibited by TIM but not PER (compare lanes 2 and 3). (B) Mixtures containing 10 fmol of dCLOCK and 10 fmol of CYC (lane 1) were incubated with decreasing amounts of PER (lanes 2 to 4), TIM (lanes 5 to 7), or PER and TIM (lanes 8 to 11). PER and TIM were added at a final concentration of 20 fmol (1×; lanes 2, 5, and 8), 4 fmol (0.2×; lanes 3, 6, and 9), 2 fmol (0.1×; lane 10), or 1 fmol (0.05×; lane 4, 7, and 11). (C) The relative intensities of the C1 complex in panel B were pooled with data obtained from two other independent experiments. The intensity of the C1 complex in the control reaction (e.g., panel B, lane 1) was set to 100, and all other values were normalized to this. “Relative molar concentrations” refers to the amount of PER and TIM in the mixture where 20 fmol is defined as 1×.
FIG. 3
PER and TIM do not inhibit the DNA binding activity of the non-bHLH-PAS transcription factor c-Rel. Reticulocyte lysates containing 7 fmol of c-Rel (lanes 3 to 5), 14 fmol of IκBα (lane 4 and 6), or 14 fmol each of PER and TIM (lane 5) were incubated with a 32P-radiolabeled synthetic oligonucleotide containing NF-κB DNA binding sites (lanes 1 to 6), and protein-DNA complexes visualized as described in the legend to Fig. 1. The positions of a complex formed by a c-Rel homodimer (top arrowhead) and an endogenously produced complex (bottom arrowhead) that is also observed when using nonprogrammed reticulocyte lysate (lane 2) are indicated.
FIG. 4
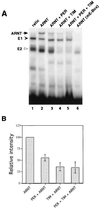
PER and TIM can inhibit the DNA binding activity of ARNT. (A) Reticulocyte lysates containing equivalent molar amounts (i.e., 20 fmol) of different exogenously produced proteins (indicated at the top) were incubated with either a wild-type (lanes 1 to 5) or mutant (lane 6) E-box [32P]DNA probe and analyzed as described in the legend to Fig. 1. A novel band is visible in the presence of ARNT (lane 2; left arrow) compared to nonprogrammed lysates (lane 1). (B) The relative intensities of the C1 complex in panel A were pooled with data obtained from two other independent experiments. The intensity of the C1 complex in the control reaction (e.g., panel A, lane 2) was set to 100, and all other values were normalized to this.
FIG. 5
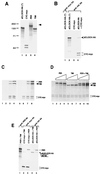
The dCLOCK-CYC complex is not disrupted by interactions with PER and/or TIM. (A) Expression plasmids containing open reading frames for different proteins (indicated at the top) were used to program a coupled transcription-translation system in the presence of [35S]methionine. An aliquot of each mixture containing 3 fmol of each target radiolabeled protein was resolved by SDS-polyacrylamide gel electrophoresis (10% polyacrylamide) and visualized by autoradiography. The positions of molecular weight standards (Bio-Rad) (in thousands) are shown at the left. A higher-molecular-weight band was routinely detected in translation reaction mixtures containing per plasmid (lane 3; identified by the right arrowhead). The production of two electrophoretic mobility variants of CYC-myc (lane 2) is due to the occasional utilization of an in-frame translation start site (data not shown). (B) Reticulocyte lysates containing 20 fmol each of [35S]methionine-labeled dCLOCK-HA (lanes 1 and 2), CYC-myc (lane 3), or both (lane 4) were incubated with antibodies (Ab) against myc (lane 1) or HA (lanes 2 to 4), and immune complexes were recovered by centrifugation. dCLOCK-HA(∗) refers to a transcription-translation reaction mixture containing no exogenously added cold (i.e., nonradiolabeled) methionine, whereas all other dCLOCK-HA proteins were produced with a mixture of [35S]methionine to nonradiolabeled methionine (1:50). The positions of dCLOCK-HA and CYC-myc are indicated on the right. (C and D) Three independent experiments are shown (C, lanes 1 to 4 and lanes 5 to 8, and D, lanes 1 to 10). Reticulocyte lysates containing 20 fmol of dCLOCK-HA (produced by using a mixture of [35S]methionine to nonradiolabeled methionine [1:400]) (as a control experiment dCLOCK-HA was omitted; C, lanes 1 and 2), 20 fmol of CYC-myc, and different amounts of PER and TIM were mixed, and immune complexes were recovered in the presence of either anti-HA antibodies (C, lanes 1 to 4 and 6 to 8, and D, lanes 1 to 10) or a nonspecific monoclonal antibody (C, lane 5). (C) The final amounts of PER and TIM in the mixtures were 20 fmol (lane 1), 60 fmol (lane 2), 20 fmol (lane 3), 60 fmol (lane 4), 60 fmol (lane 5), 0 fmol (lane 6), 20 fmol (lane 7), and 60 fmol (lane 8). (D) The final amounts of PER and TIM in the mixtures were 20 fmol (lanes 2, 5, and 8), 40 fmol (lanes 3, 6, and 9), and 60 fmol (lanes 4, 7, and 10). The positions of PER, TIM, and CYC-myc are indicated (note that because of the low-specific-activity radiolabeling, dCLOCK-HA is barely visible at the exposures shown). (E) Different combinations of in vitro-translated proteins (in these experiments dCLOCK-HA was produced with a mixture of [35S]methionine to nonradiolabeled methionine [1:50]) were subjected to immunoprecipitation with anti-PER (lane 1), anti-myc (lanes 2 and 3), or anti-HA (lane 4 and 5) antibodies and analyzed by SDS-polyacrylamide gel electrophoresis (10% polyacrylamide). In the experiment shown, 20 fmol of PER or TIM and 80 fmol of dCLOCK-HA or CYC-myc were used. The positions of PER, dCLOCK-HA, TIM, and CYC-myc are indicated on the right.
Similar articles
- dCLOCK is present in limiting amounts and likely mediates daily interactions between the dCLOCK-CYC transcription factor and the PER-TIM complex.
Bae K, Lee C, Hardin PE, Edery I. Bae K, et al. J Neurosci. 2000 Mar 1;20(5):1746-53. doi: 10.1523/JNEUROSCI.20-05-01746.2000. J Neurosci. 2000. PMID: 10684876 Free PMC article. - Closing the circadian loop: CLOCK-induced transcription of its own inhibitors per and tim.
Darlington TK, Wager-Smith K, Ceriani MF, Staknis D, Gekakis N, Steeves TD, Weitz CJ, Takahashi JS, Kay SA. Darlington TK, et al. Science. 1998 Jun 5;280(5369):1599-603. doi: 10.1126/science.280.5369.1599. Science. 1998. PMID: 9616122 - Regulation of the cycling of timeless (tim) RNA.
Wang GK, Ousley A, Darlington TK, Chen D, Chen Y, Fu W, Hickman LJ, Kay SA, Sehgal A. Wang GK, et al. J Neurobiol. 2001 Jun 5;47(3):161-75. doi: 10.1002/neu.1024. J Neurobiol. 2001. PMID: 11333398 - Circadian clocks--from genes to complex behaviour.
Roenneberg T, Merrow M. Roenneberg T, et al. Reprod Nutr Dev. 1999 May-Jun;39(3):277-94. doi: 10.1051/rnd:19990301. Reprod Nutr Dev. 1999. PMID: 10420431 Review. - Insights into the molecular mechanisms of temperature compensation from the Drosophila period and timeless mutants.
Price JL. Price JL. Chronobiol Int. 1997 Sep;14(5):455-68. doi: 10.3109/07420529709001468. Chronobiol Int. 1997. PMID: 9298282 Review.
Cited by
- Integration of light and temperature in the regulation of circadian gene expression in Drosophila.
Boothroyd CE, Wijnen H, Naef F, Saez L, Young MW. Boothroyd CE, et al. PLoS Genet. 2007 Apr 6;3(4):e54. doi: 10.1371/journal.pgen.0030054. PLoS Genet. 2007. PMID: 17411344 Free PMC article. - A role for the PERIOD:PERIOD homodimer in the Drosophila circadian clock.
Landskron J, Chen KF, Wolf E, Stanewsky R. Landskron J, et al. PLoS Biol. 2009 Apr 28;7(4):e3. doi: 10.1371/journal.pbio.1000003. PLoS Biol. 2009. PMID: 19402744 Free PMC article. - Caenorhabditis elegans period homolog lin-42 regulates the timing of heterochronic miRNA expression.
McCulloch KA, Rougvie AE. McCulloch KA, et al. Proc Natl Acad Sci U S A. 2014 Oct 28;111(43):15450-5. doi: 10.1073/pnas.1414856111. Epub 2014 Oct 15. Proc Natl Acad Sci U S A. 2014. PMID: 25319259 Free PMC article. - DOUBLETIME plays a noncatalytic role to mediate CLOCK phosphorylation and repress CLOCK-dependent transcription within the Drosophila circadian clock.
Yu W, Zheng H, Price JL, Hardin PE. Yu W, et al. Mol Cell Biol. 2009 Mar;29(6):1452-8. doi: 10.1128/MCB.01777-08. Epub 2009 Jan 12. Mol Cell Biol. 2009. PMID: 19139270 Free PMC article. - Death of a Protein: The Role of E3 Ubiquitin Ligases in Circadian Rhythms of Mice and Flies.
Abdalla OHMH, Mascarenhas B, Cheng HM. Abdalla OHMH, et al. Int J Mol Sci. 2022 Sep 12;23(18):10569. doi: 10.3390/ijms231810569. Int J Mol Sci. 2022. PMID: 36142478 Free PMC article. Review.
References
- Allada R, White N E, So W V, Hall J C, Rosbash M. A mutant Drosophila homolog of mammalian Clock disrupts circadian rhythms and transcription of period and timeless. Cell. 1998;93:791–804. - PubMed
- Baldwin A S., Jr The NF-kappa B and I kappa B proteins: new discoveries and insights. Annu Rev Immunol. 1996;14:649–683. - PubMed
- Chen Y, Hunter-Ensor M, Schotland P, Sehgal A. Alterations of per RNA in noncoding regions affect periodicity of circadian behavioral rhythms. J Biol Rhythms. 1998;13:364–379. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases