Transcription factor TFIIH is required for promoter melting in vivo - PubMed (original) (raw)
Transcription factor TFIIH is required for promoter melting in vivo
E Guzmán et al. Mol Cell Biol. 1999 Aug.
Abstract
The Rad25 protein in yeast is a DNA helicase and a subunit of the general transcription factor TFIIH. While in vitro studies have led to the hypothesis that TFIIH helicase activity plays a role in promoter melting, in vivo tests are lacking. Using potassium permanganate, which preferentially modifies single-stranded DNA, we show that a temperature-sensitive rad25(ts) mutant severely reduces the normally extensive promoter melting observed in vivo on the highly expressed genes TDH2 and PDC1 and on the induced heat shock gene HSP82. Loss of promoter melting can be observed in as little as 30 s after a shift to the nonpermissive temperature and is accompanied by a dramatic reduction in transcription. These effects on the promoter are specific, since the mutation does not affect TATA box occupancy or, in the case of HSP82, recruitment of TATA-binding protein to the TATA element or that of heat shock factor to heat shock elements. Additionally, using the technique of formaldehyde cross-linking coupled with restriction endonuclease cleavage and ligation-mediated PCR, we were able to map the polymerase density on the promoter of HSP82. This high-resolution mapping allowed us to determine that the polymerase II (Pol II) density on the promoter is also dramatically reduced after inactivation of TFIIH. These data provide strong support for the hypothesis that TFIIH functions with Pol II in the transcriptionally required step of promoter melting and show, surprisingly, that the extent of TFIIH-dependent promoter melting observed in vivo is several times larger than that seen in vitro.
Figures
FIG. 1
Primer extension of ACT1, TDH2, and PDC1 mRNAs from total RNA. Total RNA was isolated from exponentially growing yeast cultures at 25°C or after the culture had been shifted to 37°C for 2 h. The amount of total RNA was quantified, and equal amounts (30 μg) of RNA were used for primer extension with 32P end-labeled primers for ACT1, TDH2, and PDC1.
FIG. 2
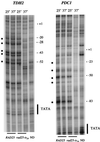
Potassium permanganate (KMnO4) reactivity of the TDH2 and PDC1 promoters in vivo. In vivo KMnO4 patterns at permissive (25°C) and nonpermissive (37°C) temperatures for both wild-type and rad25ts mutant cells are shown. The sites of cleavage were viewed by LMPCR with primers to display the bottom (transcribed) strand. The TATA sequence is labeled on the side of the gel, and the numbers indicate positions relative to the transcription start site. Permanganate-sensitive bands are labeled with bullets (•); naked DNA samples are labeled ND.
FIG. 3
Primer extension of HSP82 mRNA from total RNA. Total RNA was isolated from exponentially growing yeast cultures at 25°C or after the culture had been shifted to 39°C for 30 min. The amount of total RNA was quantified, and equal amounts (30 μg) of RNA were used for primer extension with 32P end-labeled primers for both ACT1 and HSP82. The left panel shows primer extension reactions with an actin-specific primer and a primer specific for the 5′ end of HSP82. Actin was used as an internal control due to its long half-life in vivo. The right panel shows a primer extension reaction with a primer specific for the 3′ end of HSP82. NHS, RNA extracted from yeast under non-heat shock conditions; HS, RNA extracted from yeast under heat shock conditions.
FIG. 4
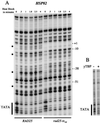
Potassium permanganate (KMnO4) reactivity of the HSP82 promoter in vivo and in vitro. (A) In vivo KMnO4 patterns before and after heat shock for both wild-type and rad25ts mutant cells. A logarithmically growing culture was treated for 1 min with 2.2 mM KMnO4 at either 25 or 39°C. The DNA was then purified and cleaved at the modified bases. The sites of cleavage were viewed by LMPCR with primers to display the bottom (transcribed) strand. The TATA sequence is labeled on the side of the gel, and the numbers indicate positions relative to the transcription start site. Permanganate-sensitive bands are labeled with bullets (•). (B) In vitro KMnO4 patterns of naked DNA with and without added yeast TBP. The promoter region of the HSP82 gene was amplified from plasmid pMF13 by PCR, and 12 fmol of this fragment, containing the TATA sequence, was treated with 25 mM KMnO4 at 25°C for 30 s in either the presence or absence of 3 pmol of yeast TBP. The primers used to amplify the fragment were UP0.1 (GAACAGGAATAAAGCTTAATCGGAT) and LARRY82 (CAGCTTGAAATTCAAAAGTTTCACT).
FIG. 5
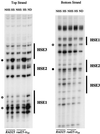
DMS footprinting of HSP82 HSEs in vivo before and after heat shock in both wild-type and rad25ts mutant cells. Four milliliters of yeast culture in exponential phase was treated with 4 μl of DMS at either 25 or 42°C for 1 min. Subsequent processing of the samples to map DMS modifications, as well as primers used to display both the top strand (left panel) and bottom strand (right panel), was performed as previously described (11). Open circles mark sites of protection, while filled circles mark sites of hypersensitivity. NHS, non-heat shock samples; HS, heat shock samples; ND, naked DNA samples.
FIG. 6
(A) Schematic representation of LMPCR. Primers specific for the HSP82 promoter are used to make blunt ends of restriction enzyme-cut DNA. Linkers are then annealed to the blunt-ended DNAs. The DNA is amplified with the original primer used to make the blunt end and one of the primers used to make the linker. The DNA is labeled with a 32P end-labeled primer which is internal to the first _HSP82_-specific primer. The products are then run on an 8.5% denaturing gel. (B) In vivo Pol II cross-linking to the HSP82 promoter in yeast cells before and after heat shock in both wild-type and rad25ts mutant cells. Yeast cells were treated with formaldehyde for 7 min either at 25°C or after a 5-min incubation at 39°C. The DNA was cut with the restriction enzymes _Mbo_II and _Hin_fI to separate the promoter from the transcribed portion of the HSP82 gene. In addition to cross-linked samples, there are also samples that were not subjected to formaldehyde cross-linking that act as a background control (NXL). After restriction cutting and immunoprecipitation steps, isolated DNA was amplified by LMPCR. The two signals amplified were judged to be from DNA that had been cut by _Mbo_II (lower band) and _Hin_fI (upper band). The percentages of total DNA cross-linked to RNA Pol II for each restriction fragment and each condition in the experiment shown are as follows: wild type non-heat shock, _Mbo_II 0.007, _Hin_fI 0.024; wild type heat shock, _Mbo_II 0.276, _Hin_fI 2.248; rad25ts non-heat shock, _Mbo_II 0.039, _Hin_fI 0.134; rad25ts heat shock, _Mbo_II 0.018, _Hin_fI 0.138.
Similar articles
- Rad25p, a DNA helicase subunit of yeast transcription factor TFIIH, is required for promoter escape in vivo.
Ostapenko D, Gileadi O. Ostapenko D, et al. Gene. 2000 Mar 7;245(1):109-17. doi: 10.1016/s0378-1119(00)00029-9. Gene. 2000. PMID: 10713451 - The FACT complex travels with elongating RNA polymerase II and is important for the fidelity of transcriptional initiation in vivo.
Mason PB, Struhl K. Mason PB, et al. Mol Cell Biol. 2003 Nov;23(22):8323-33. doi: 10.1128/MCB.23.22.8323-8333.2003. Mol Cell Biol. 2003. PMID: 14585989 Free PMC article. - Transcription-coupled DNA repair in yeast transcription factor IIE (TFIIE) mutants.
Lommel L, Gregory SM, Becker KI, Sweder KS. Lommel L, et al. Nucleic Acids Res. 2000 Feb 1;28(3):835-42. doi: 10.1093/nar/28.3.835. Nucleic Acids Res. 2000. PMID: 10637337 Free PMC article. - Promoter function and in situ protein/DNA interactions upstream of the yeast HSP90 heat shock genes.
Gross DS, Adams CC, English KE, Collins KW, Lee S. Gross DS, et al. Antonie Van Leeuwenhoek. 1990 Oct;58(3):175-86. doi: 10.1007/BF00548930. Antonie Van Leeuwenhoek. 1990. PMID: 2256678 Review. - Ten years of TFIIH.
Coin F, Egly JM. Coin F, et al. Cold Spring Harb Symp Quant Biol. 1998;63:105-10. doi: 10.1101/sqb.1998.63.105. Cold Spring Harb Symp Quant Biol. 1998. PMID: 10384274 Review. No abstract available.
Cited by
- Transcription factors that influence RNA polymerases I and II: To what extent is mechanism of action conserved?
Zhang Y, Najmi SM, Schneider DA. Zhang Y, et al. Biochim Biophys Acta Gene Regul Mech. 2017 Feb;1860(2):246-255. doi: 10.1016/j.bbagrm.2016.10.010. Epub 2016 Oct 27. Biochim Biophys Acta Gene Regul Mech. 2017. PMID: 27989933 Free PMC article. Review. - XPB induces C1D expression to counteract UV-induced apoptosis.
Li G, Liu J, Abu-Asab M, Masabumi S, Maru Y. Li G, et al. Mol Cancer Res. 2010 Jun;8(6):885-95. doi: 10.1158/1541-7786.MCR-09-0467. Epub 2010 Jun 8. Mol Cancer Res. 2010. PMID: 20530579 Free PMC article. - Role of integrative structural biology in understanding transcriptional initiation.
Trnka MJ, Pellarin R, Robinson PJ. Trnka MJ, et al. Methods. 2019 Apr 15;159-160:4-22. doi: 10.1016/j.ymeth.2019.03.009. Epub 2019 Mar 16. Methods. 2019. PMID: 30890443 Free PMC article. Review. - Genome-wide uniformity of human 'open' pre-initiation complexes.
Lai WK, Pugh BF. Lai WK, et al. Genome Res. 2017 Jan;27(1):15-26. doi: 10.1101/gr.210955.116. Epub 2016 Nov 10. Genome Res. 2017. PMID: 27927716 Free PMC article. - RNA polymerase II pausing as a context-dependent reader of the genome.
Scheidegger A, Nechaev S. Scheidegger A, et al. Biochem Cell Biol. 2016 Feb;94(1):82-92. doi: 10.1139/bcb-2015-0045. Epub 2015 Sep 15. Biochem Cell Biol. 2016. PMID: 26555214 Free PMC article. Review.
References
- Akoulitchev S, Makela T P, Weinberg R A, Reinberg D. Requirement for TFIIH kinase activity in transcription by RNA polymerase II. Nature. 1995;377:557–560. - PubMed
- Aparicio O M, Weinstein D M, Bell S P. Components and dynamics of DNA replication complexes in S. cerevisiae: redistribution of MCM proteins and Cdc45p during S phase. Cell. 1997;91:59–69. - PubMed
- Asturias F J, Meredith G D, Poglitsch C L, Kornberg R D. Two conformations of RNA polymerase II revealed by electron crystallography. J Mol Biol. 1997;272:536–540. - PubMed
- Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: Wiley Interscience; 1997.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials