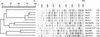Genomic relatedness of Chlamydia isolates determined by amplified fragment length polymorphism analysis - PubMed (original) (raw)
Genomic relatedness of Chlamydia isolates determined by amplified fragment length polymorphism analysis
A Meijer et al. J Bacteriol. 1999 Aug.
Abstract
The genomic relatedness of 19 Chlamydia pneumoniae isolates (17 from respiratory origin and 2 from atherosclerotic origin), 21 Chlamydia trachomatis isolates (all serovars from the human biovar, an isolate from the mouse biovar, and a porcine isolate), 6 Chlamydia psittaci isolates (5 avian isolates and 1 feline isolate), and 1 Chlamydia pecorum isolate was studied by analyzing genomic amplified fragment length polymorphism (AFLP) fingerprints. The AFLP procedure was adapted from a previously developed method for characterization of clinical C. trachomatis isolates. The fingerprints of all C. pneumoniae isolates were nearly identical, clustering together at a Dice similarity of 92.6% (+/- 1.6% standard deviation). The fingerprints of the C. trachomatis isolates of human, mouse, and swine origin were clearly distinct from each other. The fingerprints of the isolates from the human biovar could be divided into at least 12 different types when the presence or absence of specific bands was taken into account. The C. psittaci fingerprints could be divided into a parakeet, a pigeon, and a feline type. The fingerprint of C. pecorum was clearly distinct from all others. Cluster analysis of selected isolates from all species revealed groups other than those based on sequence data from single genes (in particular, omp1 and rRNA genes) but was in agreement with available DNA-DNA hybridization data. In conclusion, cluster analysis of AFLP fingerprints of representatives of all species provided suggestions for a grouping of chlamydiae based on the analysis of the whole genome. Furthermore, genomic AFLP analysis showed that the genome of C. pneumoniae is highly conserved and that no differences exist between isolates of respiratory and atherosclerotic origins.
Figures
FIG. 1
Digitized AFLP fingerprints and phylogram of 19 C. pneumoniae isolates. The phylogram was inferred by the UPGMA method with a band-based Dice similarity coefficient (S D) matrix. S D values (in percentages) and molecular sizes (Mw) (in base pairs) are shown above the phylogram and fingerprints, respectively. The names of the isolates as presented in Table 1 are shown on the right.
FIG. 2
Digitized AFLP fingerprints and phylogram of 21 C. trachomatis isolates. The phylogram was inferred by the UPGMA method with a band-based Dice similarity coefficient (S D) matrix. S D values (in percentages) and molecular sizes (Mw) (in base pairs) are shown above the phylogram and fingerprints, respectively. The names of the isolates as presented in Table 1 are shown on the right.
FIG. 3
Digitized AFLP fingerprints and phylogram of representative isolates of all four Chlamydia species. The phylogram was inferred by the UPGMA method with a band-based Dice similarity coefficient (S D) matrix. S D values (in percentages) and molecular sizes (Mw) (in base pairs) are shown above the phylogram and fingerprints, respectively. The names of the isolates as presented in Table 1 are shown on the right, with prefixes indicating the hosts (Bo, bovine; Fe, feline; Hu, human; Mu, murine; Pgn, pigeon; Prk, parakeet; Sw, swine) and species codes (Cpe, C. pecorum; Cpn, C. pneumoniae; Cps, C. psittaci; Ctr, C. trachomatis).
FIG. 4
Phylogram inferred by the UPGMA method with the DNA-DNA hybridization data matrix in Table 3. The relatedness between C. pneumoniae and C. trachomatis (murine) is missing and has been estimated as minimal (≤5%).
Similar articles
- Evolutionary relationships among members of the genus Chlamydia based on 16S ribosomal DNA analysis.
Pettersson B, Andersson A, Leitner T, Olsvik O, Uhlén M, Storey C, Black CM. Pettersson B, et al. J Bacteriol. 1997 Jul;179(13):4195-205. doi: 10.1128/jb.179.13.4195-4205.1997. J Bacteriol. 1997. PMID: 9209033 Free PMC article. - Touchdown enzyme time release-PCR for detection and identification of Chlamydia trachomatis, C. pneumoniae, and C. psittaci using the 16S and 16S-23S spacer rRNA genes.
Madico G, Quinn TC, Boman J, Gaydos CA. Madico G, et al. J Clin Microbiol. 2000 Mar;38(3):1085-93. doi: 10.1128/JCM.38.3.1085-1093.2000. J Clin Microbiol. 2000. PMID: 10699002 Free PMC article. - Detection and differentiation of Chlamydia trachomatis, Chlamydia psittaci, and Chlamydia pneumoniae by DNA amplification.
Holland SM, Gaydos CA, Quinn TC. Holland SM, et al. J Infect Dis. 1990 Oct;162(4):984-7. doi: 10.1093/infdis/162.4.984. J Infect Dis. 1990. PMID: 2401796 - Chlamydial disease--more than just abortion.
Jones GE. Jones GE. Vet J. 1997 May;153(3):249-51. doi: 10.1016/s1090-0233(97)80060-3. Vet J. 1997. PMID: 9232115 Review. No abstract available. - Chlamydial infections of the heart.
Odeh M, Oliven A. Odeh M, et al. Eur J Clin Microbiol Infect Dis. 1992 Oct;11(10):885-93. doi: 10.1007/BF01962368. Eur J Clin Microbiol Infect Dis. 1992. PMID: 1486883 Review.
Cited by
- Genome sequences of Chlamydia trachomatis MoPn and Chlamydia pneumoniae AR39.
Read TD, Brunham RC, Shen C, Gill SR, Heidelberg JF, White O, Hickey EK, Peterson J, Utterback T, Berry K, Bass S, Linher K, Weidman J, Khouri H, Craven B, Bowman C, Dodson R, Gwinn M, Nelson W, DeBoy R, Kolonay J, McClarty G, Salzberg SL, Eisen J, Fraser CM. Read TD, et al. Nucleic Acids Res. 2000 Mar 15;28(6):1397-406. doi: 10.1093/nar/28.6.1397. Nucleic Acids Res. 2000. PMID: 10684935 Free PMC article. - Comparison of whole genome sequences of Chlamydia pneumoniae J138 from Japan and CWL029 from USA.
Shirai M, Hirakawa H, Kimoto M, Tabuchi M, Kishi F, Ouchi K, Shiba T, Ishii K, Hattori M, Kuhara S, Nakazawa T. Shirai M, et al. Nucleic Acids Res. 2000 Jun 15;28(12):2311-4. doi: 10.1093/nar/28.12.2311. Nucleic Acids Res. 2000. PMID: 10871362 Free PMC article. - Chlamydial infections in urology.
Wagenlehner FM, Weidner W, Naber KG. Wagenlehner FM, et al. World J Urol. 2006 Feb;24(1):4-12. doi: 10.1007/s00345-005-0047-x. Epub 2006 Jan 19. World J Urol. 2006. PMID: 16421732 Review. - Amplified-fragment length polymorphism analysis: the state of an art.
Savelkoul PH, Aarts HJ, de Haas J, Dijkshoorn L, Duim B, Otsen M, Rademaker JL, Schouls L, Lenstra JA. Savelkoul PH, et al. J Clin Microbiol. 1999 Oct;37(10):3083-91. doi: 10.1128/JCM.37.10.3083-3091.1999. J Clin Microbiol. 1999. PMID: 10488158 Free PMC article. Review. No abstract available. - Intrastrain and interstrain genetic variation within a paralogous gene family in Chlamydia pneumoniae.
Viratyosin W, Campbell LA, Kuo CC, Rockey DD. Viratyosin W, et al. BMC Microbiol. 2002 Dec 2;2:38. doi: 10.1186/1471-2180-2-38. Epub 2002 Dec 2. BMC Microbiol. 2002. PMID: 12460455 Free PMC article.
References
- Balin B J, Gérard H C, Arking E J, Appelt D M, Branigan P J, Abrams J T, Whittum-Hudson J A, Hudson A P. Identification and localization of Chlamydia pneumoniae in the Alzheimer’s brain. Med Microbiol Immunol. 1998;87:23–42. - PubMed
- Black C M, Tharpe J A, Russell H. Distinguishing Chlamydia species by restriction analysis of the major outer membrane protein. 1992. Mol Cell Probes. 1992;6:395–400. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical