Culture-independent identification of microorganisms that respond to specified stimuli - PubMed (original) (raw)
Comparative Study
Culture-independent identification of microorganisms that respond to specified stimuli
J Borneman. Appl Environ Microbiol. 1999 Aug.
Abstract
A new approach that permits culture-independent identification of microorganisms that respond to specified stimuli was developed. This approach was illustrated by examination of microorganisms that grew in response to various nutrient supplements added to soil. A thymidine nucleotide analog, bromodeoxyuridine (BrdU), and supplements were added to soil and incubated for 3 days. DNA was extracted from the soil, and the newly synthesized DNA was isolated by immunocapture of the BrdU-labeled DNA. The unique perspective this approach offers was demonstrated by comparing the microbial community structures obtained from total soil DNA and the BrdU-labeled fraction in an rRNA gene (rDNA) analysis. The traditional total DNA analysis revealed no notable differences between the treatments, whereas the BrdU-labeled DNA showed significantly different banding patterns between the nutrient supplement treatments and compared with total DNA banding patterns. PCR primers were developed to specifically amplify the intergenic region of an rDNA sequence unique to the BrdU analysis of a phosphate supplement treatment. Amplification of DNA from all treatments using these primers showed that it was unique to the phosphate treatment and that it was present in both the total DNA and BrdU-labeled DNA fractions. This result demonstrates the promise of this new strategy, because it was able to permit identification of a sequence from a phosphate-responsive organism that was not discernable in the traditional total DNA community structure analysis.
Figures
FIG. 1
Outline of the strategy to identify microorganisms that respond to stimuli.
FIG. 2
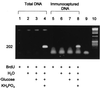
Comparison of the microbial community structures obtained from total DNA and BrdU-labeled DNA by analysis of the 16S-23S rRNA intergenic region after exposure to nutrient supplements. Lanes: 1, φX174/_Hae_III molecular weight marker (Promega); 2 and 6, treatment A; 3 and 7, treatment B; 4 and 8, treatment C; 5 and 9, treatment D.
FIG. 3
Validation of a specific rRNA gene sequence from a phosphate-responsive organism by PCR. Lanes: 9, no-template negative control; 10, 1-kb ladder (Gibco BRL, Grand Island, N.Y.).
References
- Bassam B J, Caetano-Anolles G, Gresshoff P M. Fast and sensitive staining of DNA in polyacrylamide gels. Anal Biochem. 1991;196:80–83. - PubMed
- Begg A C, McNally N J, Shrieve D C, Karcher H. A method to measure the duration of DNA synthesis and the potential doubling time from a single sample. Cytometry. 1985;6:620–626. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources