Allele-specific activators and inhibitors for kinesin - PubMed (original) (raw)
Allele-specific activators and inhibitors for kinesin
T M Kapoor et al. Proc Natl Acad Sci U S A. 1999.
Abstract
Members of the kinesin superfamily are force-generating ATPases that drive movement and influence cytoskeleton organization in cells. Often, more than one kinesin is implicated in a cellular process, and many kinesins are proposed to have overlapping functions. By using conventional kinesin as a model system, we have developed an approach to activate or inhibit a specific kinesin allele in the presence of other similar motor proteins. Modified ATP analogs are described that do not activate either conventional kinesin or another superfamily member, Eg5. However, a kinesin allele with Arg-14 in its nucleotide binding pocket mutated to alanine can use a subset of these nucleotide analogs to drive microtubule gliding. Cyclopentyl-ATP is one such analog. Cyclopentyl-adenylylimidodiphosphate, a nonhydrolyzable form of this analog, inhibits the mutant allele in microtubule-gliding assays, but not wild-type kinesin or Eg5. We anticipate that the incorporation of kinesin mutants and allele-specific activators and inhibitors in in vitro assays should clarify the role of individual motor proteins in complex cellular processes.
Figures
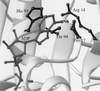
Figure 1
The nucleotide binding pocket of human conventional kinesin bound to ADP. Side chains of residues within 6 Å of the N6 nitrogen of the nucleotide are highlighted. The figure was created with
ribbons
(41).
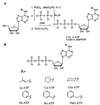
Figure 2
(A) Synthesis of the ATP and AMPPNP analogs. (B) Structures of the nucleotide analogs synthesized.
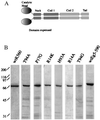
Figure 3
(A) The domain structure of the kinesin dimer. Kinesin residues including the head, neck, and a portion of the coiled-coil regions were expressed in bacteria and purified. (B) Coomasie-stained gels of the wild-type and mutant kinesins used in our assays. As assessed by gel filtration chromatography, these proteins eluted as soluble dimers.
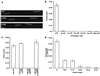
Figure 4
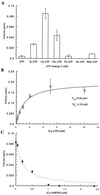
(A) Three frames from a time-lapse fluorescence microscopy video of a kinesin-driven microtubule-gliding assay are shown. The distance rhodamine-labeled microtubules move in fixed time intervals determines the velocity. The images were acquired with a 60×, 1.4 NA Nikon objective and a Princeton Instruments cooled charge-coupled device camera. Bar = 2 μm. (B) The microtubule-gliding velocity of the wild-type kinesin in the presence of different ATP analogs (1 mM) is shown. Modification of the nucleotide with a Cp group yields an ATP analog that is incapable of driving kinesin-dependent movement. (C) Cp-ATP does not inhibit wild-type kinesin motility in the presence of ATP. AMPPNP at 3.5 mM completely inhibits kinesin motility in the presence of 2 mM ATP. The nonhydrolyzable Cp-AMPPNP does not reduce ATP-dependent kinesin motility. (D) A comparison of the gliding velocities of the different mutant kinesins in the presence of 1 mM ATP.
Figure 5
Characterization of the R14A kinesin mutant. (A) Measurement of microtubule-gliding velocities in the presence of different nucleotide analogs reveals that the mutant kinesin can use Cp-ATP more efficiently than unmodified ATP. (B) R14A kinesin-driven gliding velocities depend on the Cp-ATP concentration, and the data fit well to the Michaelis Menten model [velocity = _V_max∗[Cp-ATP]/(_K_m + [Cp-ATP])]. (C) Inhibition of R14A kinesin-dependent microtubule gliding by Cp-AMPPNP, in the presence of 1 mM Cp-ATP, shows a deviation from competitive inhibition at higher concentrations (broken line) and concentrations greater than 1 mM Cp-AMPPNP completely inhibit the mutant motor. This aspect of kinesin inhibition by AMPPNP has been characterized previously (27).
Figure 6
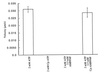
The specificity of the activator and the inhibitor for the R14A kinesin mutant. The kinesin superfamily member, Eg5, shows no detectable microtubule gliding in the presence of 2 mM Cp-ATP. Cp-AMPPNP also is unable to inhibit the enzyme at concentrations that unmodified AMPPNP completely inhibits its activity.
Similar articles
- An ATP gate controls tubulin binding by the tethered head of kinesin-1.
Alonso MC, Drummond DR, Kain S, Hoeng J, Amos L, Cross RA. Alonso MC, et al. Science. 2007 Apr 6;316(5821):120-3. doi: 10.1126/science.1136985. Science. 2007. PMID: 17412962 Free PMC article. - Coupled chemical and mechanical reaction steps in a processive Neurospora kinesin.
Crevel I, Carter N, Schliwa M, Cross R. Crevel I, et al. EMBO J. 1999 Nov 1;18(21):5863-72. doi: 10.1093/emboj/18.21.5863. EMBO J. 1999. PMID: 10545098 Free PMC article. - Nucleotide-dependent single- to double-headed binding of kinesin.
Kawaguchi K, Ishiwata S. Kawaguchi K, et al. Science. 2001 Jan 26;291(5504):667-9. doi: 10.1126/science.291.5504.667. Science. 2001. PMID: 11158681 - Kinesin: what gives?
Block SM. Block SM. Cell. 1998 Apr 3;93(1):5-8. doi: 10.1016/s0092-8674(00)81138-1. Cell. 1998. PMID: 9546384 Review. No abstract available. - Molecular motors: Kinesin's string variable.
Cross RA. Cross RA. Curr Biol. 2001 Feb 20;11(4):R147-9. doi: 10.1016/s0960-9822(01)00064-1. Curr Biol. 2001. PMID: 11250171 Review.
Cited by
- The Bump-and-Hole Tactic: Expanding the Scope of Chemical Genetics.
Islam K. Islam K. Cell Chem Biol. 2018 Oct 18;25(10):1171-1184. doi: 10.1016/j.chembiol.2018.07.001. Epub 2018 Aug 2. Cell Chem Biol. 2018. PMID: 30078633 Free PMC article. Review. - Engineered kinesin motor proteins amenable to small-molecule inhibition.
Engelke MF, Winding M, Yue Y, Shastry S, Teloni F, Reddy S, Blasius TL, Soppina P, Hancock WO, Gelfand VI, Verhey KJ. Engelke MF, et al. Nat Commun. 2016 Apr 5;7:11159. doi: 10.1038/ncomms11159. Nat Commun. 2016. PMID: 27045608 Free PMC article. - Microtubules regulate disassembly of epithelial apical junctions.
Ivanov AI, McCall IC, Babbin B, Samarin SN, Nusrat A, Parkos CA. Ivanov AI, et al. BMC Cell Biol. 2006 Mar 1;7:12. doi: 10.1186/1471-2121-7-12. BMC Cell Biol. 2006. PMID: 16509970 Free PMC article. - Bioorthogonal profiling of protein methylation using azido derivative of S-adenosyl-L-methionine.
Islam K, Bothwell I, Chen Y, Sengelaub C, Wang R, Deng H, Luo M. Islam K, et al. J Am Chem Soc. 2012 Apr 4;134(13):5909-15. doi: 10.1021/ja2118333. Epub 2012 Mar 26. J Am Chem Soc. 2012. PMID: 22404544 Free PMC article. - Formation of microtubule-based traps controls the sorting and concentration of vesicles to restricted sites of regenerating neurons after axotomy.
Erez H, Malkinson G, Prager-Khoutorsky M, De Zeeuw CI, Hoogenraad CC, Spira ME. Erez H, et al. J Cell Biol. 2007 Feb 12;176(4):497-507. doi: 10.1083/jcb.200607098. Epub 2007 Feb 5. J Cell Biol. 2007. PMID: 17283182 Free PMC article.
References
- Vale R D, Fletterick R J. Annu Rev Cell Dev Biol. 1997;13:745–777. - PubMed
- Desai A, Verma S, Mitchison T J, Walczak C E. Cell. 1999;96:69–78. - PubMed
- Wood K W, Sakowicz R, Goldstein L S, Cleveland D W. Cell. 1997;91:357–366. - PubMed
- Walczak C E, Mitchison T J. Cell. 1996;85:943–946. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources