Leptomycin B inactivates CRM1/exportin 1 by covalent modification at a cysteine residue in the central conserved region - PubMed (original) (raw)
Leptomycin B inactivates CRM1/exportin 1 by covalent modification at a cysteine residue in the central conserved region
N Kudo et al. Proc Natl Acad Sci U S A. 1999.
Abstract
The cellular target of leptomycin B (LMB), a nuclear export inhibitor, has been identified as CRM1 (exportin 1), an evolutionarily conserved receptor for the nuclear export signal of proteins. However, the mechanism by which LMB inhibits CRM1 still remains unclear. CRM1 in a Schizosaccharomyces pombe mutant showing extremely high resistance to LMB had a single amino acid replacement at Cys-529 with Ser. The mutant gene, named crm1-K1, conferred LMB resistance on wild-type S. pombe, and Crm1-K1 no longer bound biotinylated LMB. (1)H NMR analysis showed that LMB bound N-acetyl-L-cysteine methyl ester through a Michael-type addition, consistent with the idea that LMB binds covalently via its alpha, beta-unsaturated delta-lactone to the sulfhydryl group of Cys-529. When HeLa cells were cultured with biotinylated LMB, the only cellular protein bound covalently was CRM1. Inhibition by N-ethylmaleimide (NEM), an alkylating agent, of CRM1-mediated nuclear export probably was caused by covalent binding of the electrophilic structure in NEM to the sulfhydryl group of Cys-529, because the crm1-K1 mutant showed the normal rate for the export of Rev nuclear export signal-bearing proteins in the presence of not only LMB but also NEM. These results show that the single cysteine residue determines LMB sensitivity and is selectively alkylated by LMB, leading to CRM1 inactivation.
Figures
Figure 1
Isolation of an LMB-insensitive mutant and identification of the mutation. (A) A schematic representation of CRM1 functional domains and summary of allelic crm1 mutants and their phenotypes. An LMB-insensitive mutant FN41 (crm1-F1) was isolated by mutagenesis of AC1 (_crm1_-809). crm1-F1 contains a C529S mutation (TGT → AGT) besides E430K derived from _crm1_-809 (GAA → AAA). CRIME, a domain homologous to importin-β family proteins (
CR
M1,
im
portin-β,
e
tc.); Rev, a region in C-terminal-labeled CRM1 protected by Rev binding from protease digestion (26); MIC, minimal inhibitory concentration; Hs, Homo sapiens; Xl, Xenopus laevis (S. Khochbin, personal communication); Dm, Drosophila melanogaster; Ce, Caenorhabditis elegans; An, Aspergillus nidulans; Sc, Sac. cerevisiae; and Sp, S. pombe. Accession numbers are: D89729 (Hs), AC004423 (Dm), U64855 (Ce), AA966051 (An), D13039 (Sc), X15482 (Sp crm1+), and D16355 (Sp crm1-N1). (B) The single C529S mutation is sufficient for LMB resistance. S. pombe wild-type cells (JY266) were transformed with plasmids encoding crm1+ or various allelic mutant crm1 genes. FN41 and transformants were spread on YPD (1% yeast extract/2% polypeptone/2% glucose)-agar medium (−) or medium containing 10 μg/ml LMB and incubated for 5 days at 30°C. Wild-type cells transformed with empty vector (pAL19) also were tested (vector). (C) Chemical structures of LMB, biotinylated LMB, and an inactive LMB analog (i-LMB). (D) Cys-529 is essential for binding to LMB. 35S-labeled wild-type Crm1 (Sp), Crm1-K1 (K), and Sac. cerevisiae CRM1 (Sc) were synthesized in rabbit reticulocyte lysates (input) and subjected to the biotinylated LMB-binding assay after preincubation with various competitors. The eluates from immobilized streptavidin were analyzed by SDS/6% PAGE (biotin–LMB-bound).
Figure 2
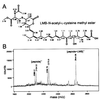
Covalent binding of leptomycin B to _N_-acetyl-
l
-cysteine methyl ester and a peptide containing a cysteine. (A) Chemical structure of an LMB complex with _N_-acetyl-
l
-cysteine methyl ester and the chemical shifts of 1H NMR. An adduct generated in the mixture containing LMB and _N_-acetyl-
l
-cysteine methyl ester was purified by HPLC, and the structure was determined by 1H NMR and correlated spectroscopy. The chemical shifts are presented with the structure. (B) An addition reaction with a peptide containing a cysteine residue. An 18-aa peptide corresponding to hCRM1 residues 513–530 that contains Cys-528 was incubated with LMB for 24 h at 37°C and analyzed by matrix-assisted laser desorption ionization–TOF MS. A peak of 2,275.9 Da may be a product of the reaction between the peptide and α-cyano-4-hydroxycinnamic acid (αCHCA) used as the matrix. Calculated molecular mass (Da) is 540 for LMB, 2,104.54 for the peptide, and 189.04 for αCHCA.
Figure 3
Nuclear export by Crm1-K1 is resistant to LMB and NEM. S. pombe strains JY266 (crm1+) and KY21 (crm1-K1) were transformed with pR1GrevNESF1 for GST-Rev NES-GFP (GST-NES-GFP) and pR1GsvNLSFrevNES1 for GST-SV40 T NLS-GFP-Rev NES (GST-NLS-GFP-NES), as described (9). These genes are under the control of the wild-type nmt1 promoter, which can strongly direct transcription when thiamin is absent in the medium. Cells were cultured in the absence of thiamin for 12 h at 30°C for induction of the proteins and then cultivated further for 6 h in the same medium with or without LMB (50 ng/ml) or NEM (100 μM) at 30°C. Cells were stained with 4′,6-diamidino-2-phenylindole for DNA and observed under a Zeiss Axiophot-2 microscope.
Figure 4
Covalent binding of biotinylated LMB to hCRM1 in vivo. HeLa cells were preincubated with 0.1% ethanol (− competitor), 100 nM LMB (LMB), or 100 nM inactive LMB analog (i-LMB) for 1 h and then treated with or without 10 nM biotinylated LMB for 2 h. After cells were lysed, the proteins bound to biotinylated LMB were isolated by using immobilized streptavidin and analyzed by electrophoresis in an SDS/5–20% polyacrylamide gradient gel followed by silver staining (Left). A single protein emerged by incubation with biotinylated LMB in the absence of LMB competitor or in the presence of inactive LMB (arrow). The proteins resolved in the polyacrylamide gel were transferred to two poly(vinylidene difluoride) membranes, one that was immunostained with a rabbit polyclonal antiserum raised against hCRM1 (Center) and another that was stained with a horseradish peroxidase-conjugated anti-biotin antibody (Right) by using a chemiluminescence system. Asterisks indicate intrinsic biotin-binding proteins such as carboxylases.
Similar articles
- The NES-Crm1p export pathway is not a major mRNA export route in Saccharomyces cerevisiae.
Neville M, Rosbash M. Neville M, et al. EMBO J. 1999 Jul 1;18(13):3746-56. doi: 10.1093/emboj/18.13.3746. EMBO J. 1999. PMID: 10393189 Free PMC article. - Leptomycin B inhibition of signal-mediated nuclear export by direct binding to CRM1.
Kudo N, Wolff B, Sekimoto T, Schreiner EP, Yoneda Y, Yanagida M, Horinouchi S, Yoshida M. Kudo N, et al. Exp Cell Res. 1998 Aug 1;242(2):540-7. doi: 10.1006/excr.1998.4136. Exp Cell Res. 1998. PMID: 9683540 - Trichostatin and leptomycin. Inhibition of histone deacetylation and signal-dependent nuclear export.
Yoshida M, Horinouchi S. Yoshida M, et al. Ann N Y Acad Sci. 1999;886:23-36. doi: 10.1111/j.1749-6632.1999.tb09397.x. Ann N Y Acad Sci. 1999. PMID: 10667200 Review. - Nucleo-cytoplasmic transport of proteins as a target for therapeutic drugs.
Yashiroda Y, Yoshida M. Yashiroda Y, et al. Curr Med Chem. 2003 May;10(9):741-8. doi: 10.2174/0929867033457791. Curr Med Chem. 2003. PMID: 12678777 Review.
Cited by
- The biology of DHX9 and its potential as a therapeutic target.
Lee T, Pelletier J. Lee T, et al. Oncotarget. 2016 Jul 5;7(27):42716-42739. doi: 10.18632/oncotarget.8446. Oncotarget. 2016. PMID: 27034008 Free PMC article. Review. - Anti-tumor activity of selective inhibitors of XPO1/CRM1-mediated nuclear export in diffuse malignant peritoneal mesothelioma: the role of survivin.
De Cesare M, Cominetti D, Doldi V, Lopergolo A, Deraco M, Gandellini P, Friedlander S, Landesman Y, Kauffman MG, Shacham S, Pennati M, Zaffaroni N. De Cesare M, et al. Oncotarget. 2015 May 30;6(15):13119-32. doi: 10.18632/oncotarget.3761. Oncotarget. 2015. PMID: 25948791 Free PMC article. - Polyglutamine Tract Expansion Increases S-Nitrosylation of Huntingtin and Ataxin-1.
Ni CL, Seth D, Fonseca FV, Wang L, Xiao TS, Gruber P, Sy MS, Stamler JS, Tartakoff AM. Ni CL, et al. PLoS One. 2016 Sep 22;11(9):e0163359. doi: 10.1371/journal.pone.0163359. eCollection 2016. PLoS One. 2016. PMID: 27658206 Free PMC article. - Inhibition of nuclear export restores nuclear localization and residual tumor suppressor function of truncated SMARCB1/INI1 protein in a molecular subset of atypical teratoid/rhabdoid tumors.
Pathak R, Zin F, Thomas C, Bens S, Gayden T, Karamchandani J, Dudley RW, Nemes K, Johann PD, Oyen F, Kordes U, Jabado N, Siebert R, Paulus W, Kool M, Frühwald MC, Albrecht S, Kalpana GV, Hasselblatt M. Pathak R, et al. Acta Neuropathol. 2021 Aug;142(2):361-374. doi: 10.1007/s00401-021-02328-w. Epub 2021 May 18. Acta Neuropathol. 2021. PMID: 34003336 Free PMC article. - Trypanosoma brucei RNA binding proteins p34 and p37 mediate NOPP44/46 cellular localization via the exportin 1 nuclear export pathway.
Hellman K, Prohaska K, Williams N. Hellman K, et al. Eukaryot Cell. 2007 Dec;6(12):2206-13. doi: 10.1128/EC.00176-07. Epub 2007 Oct 5. Eukaryot Cell. 2007. PMID: 17921352 Free PMC article.
References
- Mattaj I W, Englmeier L. Annu Rev Biochem. 1998;67:265–306. - PubMed
- Pemberton L F, Blobel G, Rosenblum J S. Curr Opin Cell Biol. 1998;10:392–399. - PubMed
- Stade K, Ford C S, Guthrie C, Weis K. Cell. 1997;90:1041–1050. - PubMed
- Fornerod M, Ohno M, Yoshida M, Mattaj I W. Cell. 1997;90:1051–1060. - PubMed
- Fukuda M, Asano S, Nakamura T, Adachi M, Yoshida M, Yanagida M, Nishida E. Nature (London) 1997;390:308–311. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases