Distinctive gene expression patterns in human mammary epithelial cells and breast cancers - PubMed (original) (raw)
. 1999 Aug 3;96(16):9212-7.
doi: 10.1073/pnas.96.16.9212.
S S Jeffrey, M van de Rijn, C A Rees, M B Eisen, D T Ross, A Pergamenschikov, C F Williams, S X Zhu, J C Lee, D Lashkari, D Shalon, P O Brown, D Botstein
Affiliations
- PMID: 10430922
- PMCID: PMC17759
- DOI: 10.1073/pnas.96.16.9212
Distinctive gene expression patterns in human mammary epithelial cells and breast cancers
C M Perou et al. Proc Natl Acad Sci U S A. 1999.
Abstract
cDNA microarrays and a clustering algorithm were used to identify patterns of gene expression in human mammary epithelial cells growing in culture and in primary human breast tumors. Clusters of coexpressed genes identified through manipulations of mammary epithelial cells in vitro also showed consistent patterns of variation in expression among breast tumor samples. By using immunohistochemistry with antibodies against proteins encoded by a particular gene in a cluster, the identity of the cell type within the tumor specimen that contributed the observed gene expression pattern could be determined. Clusters of genes with coherent expression patterns in cultured cells and in the breast tumors samples could be related to specific features of biological variation among the samples. Two such clusters were found to have patterns that correlated with variation in cell proliferation rates and with activation of the IFN-regulated signal transduction pathway, respectively. Clusters of genes expressed by stromal cells and lymphocytes in the breast tumors also were identified in this analysis. These results support the feasibility and usefulness of this systematic approach to studying variation in gene expression patterns in human cancers as a means to dissect and classify solid tumors.
Figures
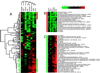
Figure 1
(A) Cluster diagram of HMEC in vitro experiments. Each column represents a single experiment, and each row represents a single gene. Ratios of gene expression relative to HMEC control samples grown under standard conditions are shown. Green squares represent lower than control levels of gene expression in the experimental samples (ratios less than 1); black squares represent genes equally expressed (ratios near 1); red squares represent higher than control levels of gene expression (ratios greater than 1); gray squares indicate insufficient or missing data. The color saturation reflects the magnitude of the log/ratio [see scale at top right and Fig. 5 (see Supplemental data at
) for the full cluster diagram with all gene names]. (B) Expanded view of the subset of genes whose expression was decreased in association with reduced HMEC proliferation. (C) Expanded view of the IFN-regulated gene cluster. In many instances, multiple independent clones/cDNA representing the same gene were spotted on different locations on these microarrays, and in most cases, these copies usually clustered together, either very near each other or immediately adjacent to each other.
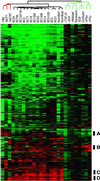
Figure 2
Overview of the combined in vitro and breast tissue specimen cluster diagram. A scaled-down representation of the 1,247-gene cluster diagram (see Supplemental Fig. 6 at
for the full cluster diagram with all gene names). The black bars show the positions of the clusters discussed in the text: (A) proliferation-associated, (B) IFN-regulated, (C) B lymphocytes, and (D) stromal cells.
Figure 3
Expanded view of two gene clusters taken from the 1,247-gene cluster diagram. (A) A portion of the proliferation-associated cluster. The numbers below each breast tumor’s column show the percentage of carcinoma cells in each specimen that stained positive for the Ki-67 antigen. (B) Expanded view of the IFN-regulated gene cluster. The letters below each breast tumor’s column identify the STAT1 staining pattern seen, with O representing no STAT1 staining (BC17 and Fig. 4_A_), W representing weak STAT1 staining, S representing strong staining (BC23 and Fig. 4_C_), P representing peripheral tumor cell nest staining (BC14 and Fig. 4_E_), and L representing staining of lymphocytes/histiocytes only (BC16 and Fig. 4_G_).
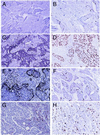
Figure 4
Immunohistochemical stains of four breast tumor specimens for the STAT1 protein (A, C, E, and G) or for the Ki-67 protein (B, D, F, and H). (A and B) Tumor BC17. (C and D) Tumor BC23. (E and F) Tumor BC14. (G and H) Tumor BC16. Magnification: approximately ×200.
References
- Ronnov-Jessen L, Petersen O W, Bissell M J. Physiol Rev. 1996;76:69–125. - PubMed
- Tavassoli F A, Schnitt S J. Pathology of the Breast. New York: Elsevier; 1992.
- DeRisi J L, Iyer V R, Brown P O. Science. 1997;278:680–686. - PubMed
- Iyer V R, Eisen M B, Ross D T, Schuler G, Moore T, Lee J C F, Trent J M, Staudt L M, Hudson J, Jr, Boguski M S, et al. Science. 1999;283:83–87. - PubMed
- Schena M, Shalon D, Davis R W, Brown P O. Science. 1995;270:467–470. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous