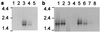Rifins: a second family of clonally variant proteins expressed on the surface of red cells infected with Plasmodium falciparum - PubMed (original) (raw)
Rifins: a second family of clonally variant proteins expressed on the surface of red cells infected with Plasmodium falciparum
S A Kyes et al. Proc Natl Acad Sci U S A. 1999.
Abstract
Many pathogens evade the host immune response or adapt to their environment by expressing surface proteins that undergo rapid switching. In the case of Plasmodium falciparum, products of a multigene family known as var are expressed on the surface of infected red cells, where they undergo clonal antigenic variation and contribute to malaria pathogenesis by mediating adhesion to a variety of host endothelial receptors and to uninfected red blood cells by forming rosettes. Herein we show that a second gene family, rif, which is associated with var at subtelomeric sites in the genome, encodes clonally variant proteins (rifins) that are expressed on the infected red cell surface. Their high copy number, sequence variability, and red cell surface location indicate an important role for rifins in malaria host-parasite interaction.
Figures
Figure 1
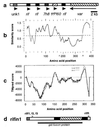
Genomic organization and sequence analysis of rifins. (a) Detail of P. falciparum chromosome 3 right subtelomeric region, showing typical orientation of rif clusters relative to subtelomeric var genes (GenBank accession no. AL010165). The clusters each contain multiple ORFs: several rif sequences; stevor/7h8, a distinct sequence with some homology to rif at the 3′ end (19); novel ORFs (e.g., unk1) of approximately 1 kb; and an apparent pseudogene (ψ) for Pf60 (35), which is highly homologous to the conserved second exon of var genes. Var is most proximal to the telomere. (b)
plotsimilarity
profile of 50 rifin sequences. Points above dotted line indicate amino acid positions of high similarity. (c) Predicted transmembrane regions and orientation relative to membrane for rifin1 predicted protein. Positions with positive
tmpred
score are possible transmembrane helices; black line indicates scores for pointing outward (i > o), gray line for pointing inward (o > i). Transmembrane regions are amino acids 3–20 (weak, pointing inward), amino acids 145–168 (strong, pointing outward), and amino acids 294–317 (strong, pointing inward). Because of length variation in the highly polymorphic region, the multiple alignment in b is longer than the single sequence in c. To align b with c, the x axis scale has been reduced in the region between arrows (∧). (d) Diagram of rifin1 predicted protein, with RT-PCR primer positions indicated (rif f1, f2, and f3 at roughly amino acid position 36–41; rifR at amino acids 318–327). Predicted features of the protein are indicated: signal (shaded), transmembrane regions (solid), semiconserved region and cytoplasmic tail ‘inside cell’ (open), polymorphic region ‘outside cell’ (hatched). Relative position of GST fusion protein is indicated by a solid line below.
Figure 2
Transcription of rif genes. Northern blot analyses of RNA, hybridized with a complex PAR+ rif probe. RNA standards are in kb. (a) Stages of PAR− parasite RNA in the erythrocytic life cycle. Lanes: 1, early rings (0–5 h after invasion); 2, middle rings (8–13 h); 3, late rings/early pigmented trophozoites (18–23 h); 4, early/mature pigmented trophozoites (24–29 h); 5, schizonts (42–47 h). (b) RNA from various parasites (late ring/early pigmented trophozoite stage): Lanes: 1, PAR+; 2, PAR−; 3, T9/96; 4, R29; 5, A4; 6, C10; 7, C18; 8, 3D7.
Figure 3
Expression of rifin proteins. (a) Western blot analysis of extracts from various parasites probed with rif3 antiserum. Lanes: 1, uninfected red cells; 2, PAR+; 3, PAR−; 4, T9/96; 5, R29; 6, A4; 7, C10; 8, C18; 9, 3D7. The 30-kDa and 46-kDa molecular mass markers are shown. (b) Immunoprecipitation of 35S metabolically labeled extracts from PAR+. Lanes: 1, rabbit preimmune serum; 2, rif1 antiserum. (c) Immunoprecipitation of extracts from 125I-surface-labeled PAR+-infected red cells. Lanes: 1 and 2, mock-trypsinized; 3 and 4, treated with trypsin at 1 mg/ml, followed by trypsin inhibitor; 1 and 3, preimmune serum; 2 and 4, rif1 antiserum. (d and e) Immunoprecipitation of triton-insoluble extracts from 35S metabolically labeled A4-infected red cells. Lanes: 1, mock-trypsinized; 2, treated with trypsin at 1 mg/ml, followed by trypsin inhibitor. (d) rif3 antiserum. (e) MESA antiserum.
Figure 4
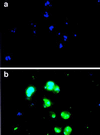
IFA of fixed PAR+-infected red cells. (a) Preimmune serum. (b) Rif3 antiserum. The secondary antibody was FITC-conjugated swine anti-rabbit immunoglobulins (green), and parasite nuclei were stained with 4,6-diamidino-2-phenylindole (blue). The rif1 antiserum gave similar results.
Figure 5
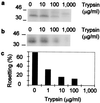
Trypsin sensitivity of rifins and rosetting in PAR+. PAR+-infected red cells were surface-labeled with 125I and then treated with various concentrations of trypsin. (a) Total Triton X-100-soluble extracts. (b) Triton X-100-soluble extracts immunoprecipitated with rif3 antiserum. Both were subjected to SDS/PAGE. (c) Rosetting of trypsinized infected cells. The percentage rosetting indicates the proportion of mature infected red cells binding two or more uninfected red cells. The data are from a representative experiment, and three further repetitions gave similar results.
Similar articles
- Small, clonally variant antigens expressed on the surface of the Plasmodium falciparum-infected erythrocyte are encoded by the rif gene family and are the target of human immune responses.
Fernandez V, Hommel M, Chen Q, Hagblom P, Wahlgren M. Fernandez V, et al. J Exp Med. 1999 Nov 15;190(10):1393-404. doi: 10.1084/jem.190.10.1393. J Exp Med. 1999. PMID: 10562315 Free PMC article. - Variant surface antigens of Plasmodium falciparum and their roles in severe malaria.
Wahlgren M, Goel S, Akhouri RR. Wahlgren M, et al. Nat Rev Microbiol. 2017 Aug;15(8):479-491. doi: 10.1038/nrmicro.2017.47. Epub 2017 Jun 12. Nat Rev Microbiol. 2017. PMID: 28603279 Review. - Multiple var gene transcripts are expressed in Plasmodium falciparum infected erythrocytes selected for adhesion.
Noviyanti R, Brown GV, Wickham ME, Duffy MF, Cowman AF, Reeder JC. Noviyanti R, et al. Mol Biochem Parasitol. 2001 May;114(2):227-37. doi: 10.1016/s0166-6851(01)00266-3. Mol Biochem Parasitol. 2001. PMID: 11378202 - Expression switching in the stevor and Pfmc-2TM superfamilies in Plasmodium falciparum.
Lavazec C, Sanyal S, Templeton TJ. Lavazec C, et al. Mol Microbiol. 2007 Jun;64(6):1621-34. doi: 10.1111/j.1365-2958.2007.05767.x. Mol Microbiol. 2007. PMID: 17555442 - [Var gene family and antigen variation in Plasmodium falciparum].
Fang XN. Fang XN. Zhongguo Ji Sheng Chong Xue Yu Ji Sheng Chong Bing Za Zhi. 2010 Apr;28(2):153-6. Zhongguo Ji Sheng Chong Xue Yu Ji Sheng Chong Bing Za Zhi. 2010. PMID: 20666324 Review. Chinese.
Cited by
- Targets of antibodies against Plasmodium falciparum-infected erythrocytes in malaria immunity.
Chan JA, Howell KB, Reiling L, Ataide R, Mackintosh CL, Fowkes FJ, Petter M, Chesson JM, Langer C, Warimwe GM, Duffy MF, Rogerson SJ, Bull PC, Cowman AF, Marsh K, Beeson JG. Chan JA, et al. J Clin Invest. 2012 Sep;122(9):3227-38. doi: 10.1172/JCI62182. Epub 2012 Aug 1. J Clin Invest. 2012. PMID: 22850879 Free PMC article. - Antigenic differences and conservation among placental Plasmodium falciparum-infected erythrocytes and acquisition of variant-specific and cross-reactive antibodies.
Beeson JG, Mann EJ, Byrne TJ, Caragounis A, Elliott SR, Brown GV, Rogerson SJ. Beeson JG, et al. J Infect Dis. 2006 Mar 1;193(5):721-30. doi: 10.1086/500145. Epub 2006 Jan 30. J Infect Dis. 2006. PMID: 16453269 Free PMC article. - Antigenic diversity and immune evasion by malaria parasites.
Ferreira MU, da Silva Nunes M, Wunderlich G. Ferreira MU, et al. Clin Diagn Lab Immunol. 2004 Nov;11(6):987-95. doi: 10.1128/CDLI.11.6.987-995.2004. Clin Diagn Lab Immunol. 2004. PMID: 15539495 Free PMC article. Review. No abstract available. - A Plasmodium gene family encoding Maurer's cleft membrane proteins: structural properties and expression profiling.
Sam-Yellowe TY, Florens L, Johnson JR, Wang T, Drazba JA, Le Roch KG, Zhou Y, Batalov S, Carucci DJ, Winzeler EA, Yates JR 3rd. Sam-Yellowe TY, et al. Genome Res. 2004 Jun;14(6):1052-9. doi: 10.1101/gr.2126104. Epub 2004 May 12. Genome Res. 2004. PMID: 15140830 Free PMC article. - Plasmodium falciparum virulence determinants unveiled.
Crabb BS, Cowman AF. Crabb BS, et al. Genome Biol. 2002 Oct 25;3(11):REVIEWS1031. doi: 10.1186/gb-2002-3-11-reviews1031. Epub 2002 Oct 25. Genome Biol. 2002. PMID: 12441004 Free PMC article. Review.
References
- Newbold C I, Craig A G, Kyes S, Berendt A R, Snow R W, Peshu N, Marsh K. Ann Trop Med Parasitol. 1997;91:551–557. - PubMed
- Kaul D K, Roth E F, Jr, Nagel R L, Howard R J, Handunnetti S M. Blood. 1991;78:812–819. - PubMed
- Warrell D A. Parasitology. 1987;94:S53–S76. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases