Generation of influenza A viruses entirely from cloned cDNAs - PubMed (original) (raw)
. 1999 Aug 3;96(16):9345-50.
doi: 10.1073/pnas.96.16.9345.
T Watanabe, H Ito, S Watanabe, H Goto, P Gao, M Hughes, D R Perez, R Donis, E Hoffmann, G Hobom, Y Kawaoka
Affiliations
- PMID: 10430945
- PMCID: PMC17785
- DOI: 10.1073/pnas.96.16.9345
Generation of influenza A viruses entirely from cloned cDNAs
G Neumann et al. Proc Natl Acad Sci U S A. 1999.
Abstract
We describe a new reverse-genetics system that allows one to efficiently generate influenza A viruses entirely from cloned cDNAs. Human embryonic kidney cells (293T) were transfected with eight plasmids, each encoding a viral RNA of the A/WSN/33 (H1N1) or A/PR/8/34 (H1N1) virus, flanked by the human RNA polymerase I promoter and the mouse RNA polymerase I terminator-together with plasmids encoding viral nucleoprotein and the PB2, PB1, and PA viral polymerases. This strategy yielded >1 x 10(3) plaque-forming units (pfu) of virus per ml of supernatant at 48 hr posttransfection. The addition of plasmids expressing all of the remaining viral structural proteins led to a substantial increase in virus production, 3 x 10(4)-5 x 10(7) pfu/ml. We also used reverse genetics to generate a reassortant virus containing the PB1 gene of the A/PR/8/34 virus, with all other genes representing A/WSN/33. Additional viruses produced by this method had mutations in the PA gene or possessed a foreign epitope in the head of the neuraminidase protein. This efficient system, which does not require helper virus infection, should be useful in viral mutagenesis studies and in the production of vaccines and gene therapy vectors.
Figures
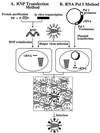
Figure 1
Schematic diagram of established reverse-genetics systems. In the RNP transfection method (A), purified NP and polymerase proteins are assembled into RNPs with the use of _in vitro_-synthesized vRNA. Cells are transfected with RNPs, followed by helper virus infection. In the RNA polymerase I method (B), a plasmid containing the RNA polymerase I promoter, a cDNA encoding the vRNA to be rescued, and the RNA polymerase I terminator are transfected into cells. Intracellular transcription by RNA polymerase I yields synthetic vRNA, which is packaged into progeny virus particles upon infection with helper virus. With both methods, transfectant viruses (i.e., those containing RNA derived from cloned cDNA) are selected from the helper virus population.
Figure 2
Schematic diagram of the generation of RNA polymerase I constructs. cDNAs derived from influenza virus were amplified by PCR, digested with _Bsm_BI, and cloned into the _Bsm_BI sites of the pHH21 vector (34), which contains the human RNA polymerase I promoter (P) and the mouse RNA polymerase I terminator (T). The thymidine nucleotide upstream of the terminator sequence (*T) represents the 3′ end of the influenza viral RNA. Influenza A virus sequences are shown in boldface letters.
Figure 3
Reverse-genetics method for generating segmented, negative-sense RNA viruses entirely from cloned cDNA. Plasmids containing the RNA polymerase I promoter, a cDNA for each of the eight viral RNA segments, and the RNA polymerase I terminator are transfected into cells together with protein expression plasmids. Although infectious viruses can be generated with plasmids expressing PA, PB1, PB2, and NP, expression of all remaining structural proteins (shown in brackets) increases the efficiency of virus production (see text).
Figure 4
Detection of the Flag epitope in cells infected with a transfectant virus. Antibody staining was used to identify the NA in MDCK cells infected with either PR8-WSN-FL79 (A and D) or A/WSN/33 wild-type virus (B and E) or on mock-infected MDCK cells (C and F). Nine hours after infection, cells were fixed with paraformaldehyde, treated with Triton X-100, and incubated with either anti-Flag (A_–_C) or anti-WSN NA (D_–_F) mAbs. Intensive Golgi staining (red) is apparent in positive samples (A, D, and E).
Figure 5
Recovery of PA mutants. The PA gene of each virus was amplified by reverse transcriptase–PCR with primers that yield a 1,226-bp fragment (position 677-1903 of the mRNA; lanes 1, 3, and 5), which then was digested with the restriction enzyme _Bsp_120I (at position 846 of the mRNA; lanes 4 and 7) or _Pvu_II (at position 1284 of the mRNA; lanes 2 and 6). The presence of _Bsp_120I or _Pvu_II sites in the PCR products yielded either 169- and 1,057-bp or 607- and 619-bp fragments, respectively. MW, molecular weight markers.
Comment in
- Reverse genetics of negative-strand RNA viruses: closing the circle.
Pekosz A, He B, Lamb RA. Pekosz A, et al. Proc Natl Acad Sci U S A. 1999 Aug 3;96(16):8804-6. doi: 10.1073/pnas.96.16.8804. Proc Natl Acad Sci U S A. 1999. PMID: 10430844 Free PMC article. Review. No abstract available.
Similar articles
- A DNA transfection system for generation of influenza A virus from eight plasmids.
Hoffmann E, Neumann G, Kawaoka Y, Hobom G, Webster RG. Hoffmann E, et al. Proc Natl Acad Sci U S A. 2000 May 23;97(11):6108-13. doi: 10.1073/pnas.100133697. Proc Natl Acad Sci U S A. 2000. PMID: 10801978 Free PMC article. - Rescue of influenza C virus from recombinant DNA.
Crescenzo-Chaigne B, van der Werf S. Crescenzo-Chaigne B, et al. J Virol. 2007 Oct;81(20):11282-9. doi: 10.1128/JVI.00910-07. Epub 2007 Aug 8. J Virol. 2007. PMID: 17686850 Free PMC article. - Efficient generation and growth of influenza virus A/PR/8/34 from eight cDNA fragments.
de Wit E, Spronken MI, Bestebroer TM, Rimmelzwaan GF, Osterhaus AD, Fouchier RA. de Wit E, et al. Virus Res. 2004 Jul;103(1-2):155-61. doi: 10.1016/j.virusres.2004.02.028. Virus Res. 2004. PMID: 15163504 - Generation of influenza A virus from cloned cDNAs--historical perspective and outlook for the new millenium.
Neumann G, Kawaoka Y. Neumann G, et al. Rev Med Virol. 2002 Jan-Feb;12(1):13-30. doi: 10.1002/rmv.332. Rev Med Virol. 2002. PMID: 11787081 Review. - Reverse genetics systems for the generation of segmented negative-sense RNA viruses entirely from cloned cDNA.
Neumann G, Kawaoka Y. Neumann G, et al. Curr Top Microbiol Immunol. 2004;283:43-60. doi: 10.1007/978-3-662-06099-5_2. Curr Top Microbiol Immunol. 2004. PMID: 15298167 Review.
Cited by
- Transmission of a 2009 H1N1 pandemic influenza virus occurs before fever is detected, in the ferret model.
Roberts KL, Shelton H, Stilwell P, Barclay WS. Roberts KL, et al. PLoS One. 2012;7(8):e43303. doi: 10.1371/journal.pone.0043303. Epub 2012 Aug 29. PLoS One. 2012. PMID: 22952661 Free PMC article. - Rescue of Infectious Salmon Anemia Virus (ISAV) from Cloned cDNA.
Toro-Ascuy D, Cárdenas M, Vásquez-Martínez Y, Cortez-San Martín M. Toro-Ascuy D, et al. Methods Mol Biol. 2024;2733:87-99. doi: 10.1007/978-1-0716-3533-9_6. Methods Mol Biol. 2024. PMID: 38064028 - A Bivalent Vaccine Based on a PB2-Knockout Influenza Virus Protects Mice From Secondary Pneumococcal Pneumonia.
Uraki R, Piao Z, Akeda Y, Iwatsuki-Horimoto K, Kiso M, Ozawa M, Oishi K, Kawaoka Y. Uraki R, et al. J Infect Dis. 2015 Dec 15;212(12):1939-48. doi: 10.1093/infdis/jiv341. Epub 2015 Jun 29. J Infect Dis. 2015. PMID: 26123562 Free PMC article. - Development of an improved polykaryon-based influenza virus rescue system.
Bourret V, Lyall J, Ducatez MF, Guérin JL, Tiley L. Bourret V, et al. BMC Biotechnol. 2012 Sep 25;12:69. doi: 10.1186/1472-6750-12-69. BMC Biotechnol. 2012. PMID: 23009349 Free PMC article. - Reverse Genetics Approaches to Control Arenavirus.
Martínez-Sobrido L, Cheng BY, de la Torre JC. Martínez-Sobrido L, et al. Methods Mol Biol. 2016;1403:313-51. doi: 10.1007/978-1-4939-3387-7_17. Methods Mol Biol. 2016. PMID: 27076139 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous