Identification of a new regulator in Streptococcus pneumoniae linking quorum sensing to competence for genetic transformation - PubMed (original) (raw)
Identification of a new regulator in Streptococcus pneumoniae linking quorum sensing to competence for genetic transformation
M S Lee et al. J Bacteriol. 1999 Aug.
Abstract
Competence for genetic transformation in Streptococcus pneumoniae is regulated by a quorum-sensing system encoded by two genetic loci, comCDE and comAB. Additional competence-specific operons, cilA, cilB, cilC, cilD, cilE, cinA-recA, coiA, and cfl, involved in the DNA uptake process and recombination, share an unusual consensus sequence at -10 and -25 in the promoter, which is absent from the promoters of comAB and comCDE. This pattern suggests that a factor regulating transcription of these transformation machinery genes but not involved with comCDE and comAB expression might be an alternative sigma factor. A search for such a global transcriptional regulator was begun by purifying pneumococcal RNA polymerase holoenzyme. In preparations from competent pneumococcal cultures a protein which seemed to be responsible for cilA transcription in vitro was identified. The corresponding gene was identified and found to be present in two copies, designated comX1 and comX2, located adjacent to two of the repeated rRNA operons. Expression of transformation machinery operons, such as cilA, cilD, cilE, and cfl, but not that of the quorum-sensing operons comAB and comCDE, was shown to depend on comX, while comX expression depended on ComE but not on ComX itself. We conclude that the factor is a competence-specific global transcription modulator which links quorum-sensing information transduced to ComE to competence and propose that it acts as an alternate sigma factor. We also report that comAB and comCDE are not sufficient for shutoff of competence-stimulating peptide-induced gene expression nor for the subsequent refractory period, suggesting that these phenomena depend on one or more ComX-dependent genes.
Figures
FIG. 1
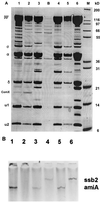
Specific transcription of a competence gene, ssb2, by preparations of His-tagged RNA polymerase with ComX. (A) SDS-PAGE analysis of RNA polymerase preparations from competent and noncompetent cultures is shown. RNA polymerase was prepared from three competent cultures (lanes 1, 2, and 3) and from three independent noncompetent cultures (lanes 4, 5, and 6). Lane B was blank (a control preparation with no His tag from CP1250), and lane M contained standards. The amounts of protein loaded were approximately 25, 10, 25, 25, 10, and 50 μg, from left to right, respectively. The gel was stained with Colloidal Blue stain. (B) Transcriptional specificity of RNA polymerase preparations. In vitro transcription of two genes, amiA and ssb2, was performed with three RNA polymerase preparations as described in the Materials and Methods section. The reaction products were analyzed on a denaturing 6% polyacrylamide gel. Products are shown for two preparations from competent cultures (lanes 3 and 4, enzyme displayed in panel A, lane 1; lanes 5 and 6, enzyme displayed in panel A, lane 2) and one from a noncompetent culture (lanes 1 and 2, enzyme displayed in panel A, lane 6). Templates were ssb2 for lanes 2, 4, and 6 and amiA for lanes 1, 3, and 5. The predicted transcript sizes are 420 bp for ssb2 and 300 bp for amiA.
FIG. 2
Map of regions neighboring comX1 and comX2. Solid vertical lines represent the ends of sequence read for each locus, and the dotted vertical line marks the border of structural identity between the loci. Open pentagons represent putative gene assignments; filled pentagons indicate comX copies. Ter represents rho-independent terminator; bent arrows represent promoters. The complete map at the top is derived from the partial type 4 genome sequence. The organization of the sequence flanking comX1 in CP1250 was the same as that of contig 4125 from type 4. The comX2 locus was absent from the type 4 genome sequence database and was determined in CP1250 as described in the text.
FIG. 3
Strategies for construction and confirmation of an insertion-deletion mutation of comX1 and a comX1::lacZ fusion. Solid arrows and arrowheads mark primers for PCR; hollow arrows represent transformation processes. Hatched pentagon represents comX1 gene and its direction of transcription. Boxes and lines indicate double-stranded DNA. Black boxes in DNA represent PcEm markers. (A) Construction of CPM2 by transformation of CP1250 with the amplicon aMSL2, assembled by PCR from an Emr cassette and two DNA fragments flanking comX1. (B) Construction of CPM3 by transformation of CP1250 with pMSL2, an insertion plasmid targeting comX1. Details of construction of the strains and the verification of marker integration are described in the Materials and Methods section.
FIG. 4
Sequence alignment of ComX homologues. The sequences of the homologues were aligned with the program Clustal W (9). Amino acid similarity is indicated by highlighting (black background, ≥50% identity; shaded background, ≥50% similarity). In the consensus sequence, residues with ≥80% similarity are shown in lowercase and 100% matches are shown in uppercase. The most highly conserved region of the sigma 70 family (subregion 2.2) is located between residues 56 and 75 of B. subtilis sigH. Protein abbreviations and sources are as follows: SPN ComX, ComX in S. pneumoniae, accession no. RPN00272 in the WIT database (52); SMU ORF, S. mutans, contig 746 (bp 2275 through 1796 of the sequence in the NCBI database [36]); SPY ORF, S. pyogenes accession no. RST00265 in the WIT database; LLA hSigH, L. lactis tr/Q48591; EFA hSigH, E. faecalis REF02274 in the WIT database; BSU SigH, B. subtilis RBS00098 in the WIT database.
FIG. 5
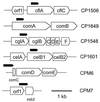
Map of lacZ fusion sites at six competence loci. Each targeting fragment (filled pentagons) directed the nonreplicative vector, pEVP3, to introduce a lacZ fusion at the site marked by the point of the pentagon. Fusion constructs except that of CPM7 (see the Methods and Materials section) were described previously (24). The exact positions of the targeting fragments in the competence loci in the mutants are as follows: strain CP1506, contig 4155 (bp 6235 through 5935); CP1649, contig 4105 (bp 4920 through 5220); CP1548, contig 4194 (bp 16895 through 16595); CP1601, contig 4139 (bp 4806 through 4506); CPM6, U33315 (bp 332 through 721); CPM7, contig 4219 (bp 836 through 476). Gene positions are as follows: cflA, contig 4155 (bp 6454 through 5156); celB1, contig 4139 (bp 5029 through 3506); comA, contig 4105 (bp 4693 through 6852); cglA, contig 4194 (bp 17495 through 16554); ssb2, contig 4219 (bp 514 through 118).
FIG. 6
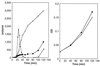
Comparison of comX promoter activities in CPM16 and CPM4 after competence induction. Competence was induced by CSP and NaOH (at time 0) for each strain at OD550 of 0.025. Left panel: β-galactosidase activities (Miller units) were monitored for both strains CPM16 (●) and strain CPM4 (○). Transformation (1 unit = 100 Novr transformants) for CPM16 (■) was also monitored at the same time by determining the number of Novr transformants after exposing a portion of the culture to donor DNA for 90 s. Right panel: Growth of CPM16 (●) and CPM4 (○) was monitored by measuring OD550.
FIG. 7
Hypothetical model for the regulation of genetic transformation. Solid arrows indicate processing steps or transcriptional activation steps that have been shown to take place or for which supporting observations are described in the text. T bars indicate negative regulation. Dashed lines indicate hypothetical links. comI, a putative gene responsible for competence shutoff and refractory period.
Similar articles
- ComX is a unique link between multiple quorum sensing outputs and competence in Streptococcus pneumoniae.
Luo P, Li H, Morrison DA. Luo P, et al. Mol Microbiol. 2003 Oct;50(2):623-33. doi: 10.1046/j.1365-2958.2003.03714.x. Mol Microbiol. 2003. PMID: 14617184 - Cross-regulation of competence pheromone production and export in the early control of transformation in Streptococcus pneumoniae.
Martin B, Prudhomme M, Alloing G, Granadel C, Claverys JP. Martin B, et al. Mol Microbiol. 2000 Nov;38(4):867-78. doi: 10.1046/j.1365-2958.2000.02187.x. Mol Microbiol. 2000. PMID: 11115120 - Regulation of competence for genetic transformation in Streptococcus pneumoniae: a link between quorum sensing and DNA processing genes.
Morrison DA, Lee MS. Morrison DA, et al. Res Microbiol. 2000 Jul-Aug;151(6):445-51. doi: 10.1016/s0923-2508(00)00171-6. Res Microbiol. 2000. PMID: 10961457 Review. - Quorum Sensing Regulation of Competence and Bacteriocins in Streptococcus pneumoniae and mutans.
Shanker E, Federle MJ. Shanker E, et al. Genes (Basel). 2017 Jan 5;8(1):15. doi: 10.3390/genes8010015. Genes (Basel). 2017. PMID: 28067778 Free PMC article. Review.
Cited by
- Cell death of Streptococcus mutans induced by a quorum-sensing peptide occurs via a conserved streptococcal autolysin.
Dufour D, Lévesque CM. Dufour D, et al. J Bacteriol. 2013 Jan;195(1):105-14. doi: 10.1128/JB.00926-12. Epub 2012 Oct 26. J Bacteriol. 2013. PMID: 23104806 Free PMC article. - Competence for natural genetic transformation in the Streptococcus bovis group streptococci S. infantarius and S. macedonicus.
Morrison DA, Guédon E, Renault P. Morrison DA, et al. J Bacteriol. 2013 Jun;195(11):2612-20. doi: 10.1128/JB.00230-13. Epub 2013 Mar 29. J Bacteriol. 2013. PMID: 23543718 Free PMC article. - hsdSA regulated extracellular vesicle-associated PLY to protect Streptococcus pneumoniae from macrophage killing via LAPosomes.
Wang L, Liu M, Qi Y, Wang J, Shi Q, Xie X, Zhou C, Ma L. Wang L, et al. Microbiol Spectr. 2024 Jan 11;12(1):e0099523. doi: 10.1128/spectrum.00995-23. Epub 2023 Nov 29. Microbiol Spectr. 2024. PMID: 38018988 Free PMC article. - Antibacterial effects of Traditional Chinese Medicine monomers against Streptococcus pneumoniae via inhibiting pneumococcal histidine kinase (VicK).
Zhang S, Wang J, Xu W, Liu Y, Wang W, Wu K, Wang Z, Zhang X. Zhang S, et al. Front Microbiol. 2015 May 20;6:479. doi: 10.3389/fmicb.2015.00479. eCollection 2015. Front Microbiol. 2015. PMID: 26042111 Free PMC article. - Programmed protection of foreign DNA from restriction allows pathogenicity island exchange during pneumococcal transformation.
Johnston C, Martin B, Granadel C, Polard P, Claverys JP. Johnston C, et al. PLoS Pathog. 2013 Feb;9(2):e1003178. doi: 10.1371/journal.ppat.1003178. Epub 2013 Feb 14. PLoS Pathog. 2013. PMID: 23459610 Free PMC article.
References
- Aaberge I S, Eng J, Lermark G, Løvik M. Virulence of Streptococcus pneumoniae in mice: a standardized method for preparation and frozen storage of the experimental bacterial inoculum. Microb Pathog. 1995;18:141–152. - PubMed
- Alloing G, Trombe M C, Claverys J P. The ami locus of the gram-positive bacterium Streptococcus pneumoniae is similar to binding protein-dependent transport operons of gram-negative bacteria. Mol Microbiol. 1990;4:633–644. - PubMed
- Alloing G, Granadel C, Morrison D A, Claverys J P. Competence pheromone, oligopeptide permease, and induction of competence in Streptococcus pneumoniae. Mol Microbiol. 1996;21:471–478. - PubMed
- Alloing G, Martin B, Granadel C, Claverys J P. Development of competence in Streptococcus pneumoniae: pheromone autoinduction and control of quorum-sensing by the oligopeptide permease. Mol Microbiol. 1998;29:75–83. - PubMed
- Campbell E A, Choi S Y, Masure H R. A competence regulon in Streptococcus pneumoniae revealed by genomic analysis. Mol Microbiol. 1998;27:929–939. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases