The speed limit for protein folding measured by triplet-triplet energy transfer - PubMed (original) (raw)
The speed limit for protein folding measured by triplet-triplet energy transfer
O Bieri et al. Proc Natl Acad Sci U S A. 1999.
Abstract
A direct measure of intramolecular chain diffusion is obtained by the determination of triplet-triplet energy-transfer rates between a donor and an acceptor chromophore attached at defined points on a polypeptide chain. Single exponential kinetics of contact formation are observed on the nanosecond time scale for polypeptides in which donor and acceptor are linked by repeating units of glycine and serine residues. The rates depend on the number of peptide bonds (N) separating donor and acceptor and show a maximum for the shortest peptides (N = 3) with a time constant (tau = 1/k) of 20 ns. This sets an upper limit for the speed of formation of the first side-chain contacts during protein folding.
Figures
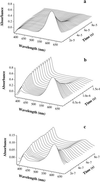
Figure 1
Transient absorbance spectra of thioxanthone (a) and a mixture of thioxanthone and naphthyl-acetic acid (b) after selective excitation of thioxanthone by a 20-ns laser pulse at 351 nm. The concentration of thioxanthone was 86 μM in both cases. The populations of donor (thioxanthone) and acceptor (1-naphthylalanine) triplets can be easily measured by their strong absorbance bands at 620 nm and 420 nm, respectively. The half-life of triplet thioxanthone in the absence of acceptor (a) was 30 μs with the experimental conditions used. The decay was of mixed order because of triplet–triplet annihilation. In the presence of 0.5 mM 1-naphthylalanine (b), the lifetime of thioxanthone triplets decreases to 500 ns. This effect is due to triplet–triplet energy transfer to the acceptor chromophore as indicated by the concomitant growth of triplet absorbance by 1-naphthylalanine at 420 nm. c displays the same experiments as b, but with donor and acceptor chromophores attached to a polypeptide chain [peptide A(n = 4)]. The experiments were carried out in ethanol. For intramolecular transfer (c), the solution contained 40% glycerol. This slows contact formation (compare Fig. 4) and thus enables the recording of time resolved spectra. The peptide concentration was 15 μM.
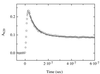
Figure 2
Transient triplet absorbance of thioxanthone after 351-nm laser flash photolysis of peptide A(n = 4) in ethanol. The laserflash was applied at t = 0. The decrease in thioxanthone triplet absorbance at 620 nm (○) can be described by a single-exponential process with a rate constant of 1.4 ± 0.2 × 107 s−1 (solid line). Because intermolecular triplet-energy transfer was shown to be diffusion-controlled, this rate corresponds to the rate of intrachain diffusion between donor and acceptor. The peptide concentration was 40 μM. Under these conditions, the half-life of thioxanthone triplets in peptides containing no acceptor was 30 μs (data not shown). The experiments were carried out in ethanol at 22°C. About 30% of the thioxanthone molecules remain in the triplet state after the equilibrium is reached, because of the small difference in triplet energy between donor and acceptor in polar solvents. These equilibrated triplet states decay on a much slower time scale and with the same rate as decay of the acceptor triplets occurs (1/k ≈30 μs; data not shown).
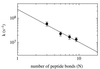
Figure 3
Distance dependence of the rate of intramolecular chain diffusion. The rates of contact formation in the different peptides A(n = 1–4) are plotted against the number of peptide bonds (N) separating donor and acceptor. Experimental conditions were as in Fig. 2. The peptide concentration was 15 μM. A linear fit of the double exponential plot gives a slope of −1.36 ± 0.26.
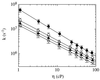
Figure 4
Viscosity dependence of the rate of contact formation for peptides A containing n = 1 (●), 2 (○), 3 (▴) and 4 (▵) glycine/serine pairs between thioxanthone and 1-naphthylalanine. Linear fits of the double logarithmic plot of the data give slopes of −0.96 ± 0.05, −0.83 ± 0.05, −0.80 ± 0.05, and −0.81 ± 0.05, respectively.
Similar articles
- Loop formation in unfolded polypeptide chains on the picoseconds to microseconds time scale.
Fierz B, Satzger H, Root C, Gilch P, Zinth W, Kiefhaber T. Fierz B, et al. Proc Natl Acad Sci U S A. 2007 Feb 13;104(7):2163-8. doi: 10.1073/pnas.0611087104. Epub 2007 Feb 6. Proc Natl Acad Sci U S A. 2007. PMID: 17284588 Free PMC article. - Dynamics of unfolded polypeptide chains as model for the earliest steps in protein folding.
Krieger F, Fierz B, Bieri O, Drewello M, Kiefhaber T. Krieger F, et al. J Mol Biol. 2003 Sep 5;332(1):265-74. doi: 10.1016/s0022-2836(03)00892-1. J Mol Biol. 2003. PMID: 12946363 - Triplet-triplet energy transfer studies on conformational dynamics in peptides and a protein.
Reiner A. Reiner A. J Pept Sci. 2011 Jun;17(6):413-9. doi: 10.1002/psc.1353. Epub 2011 Feb 24. J Pept Sci. 2011. PMID: 21360629 Review. - Measuring the rate of intramolecular contact formation in polypeptides.
Lapidus LJ, Eaton WA, Hofrichter J. Lapidus LJ, et al. Proc Natl Acad Sci U S A. 2000 Jun 20;97(13):7220-5. doi: 10.1073/pnas.97.13.7220. Proc Natl Acad Sci U S A. 2000. PMID: 10860987 Free PMC article. - Using triplet-triplet energy transfer to measure conformational dynamics in polypeptide chains.
Fierz B, Joder K, Krieger F, Kiefhaber T. Fierz B, et al. Methods Mol Biol. 2007;350:169-87. doi: 10.1385/1-59745-189-4:169. Methods Mol Biol. 2007. PMID: 16957323 Review.
Cited by
- Loop formation in unfolded polypeptide chains on the picoseconds to microseconds time scale.
Fierz B, Satzger H, Root C, Gilch P, Zinth W, Kiefhaber T. Fierz B, et al. Proc Natl Acad Sci U S A. 2007 Feb 13;104(7):2163-8. doi: 10.1073/pnas.0611087104. Epub 2007 Feb 6. Proc Natl Acad Sci U S A. 2007. PMID: 17284588 Free PMC article. - Direct quantification of the attempt frequency determining the mechanical unfolding of ubiquitin protein.
Popa I, Fernández JM, Garcia-Manyes S. Popa I, et al. J Biol Chem. 2011 Sep 9;286(36):31072-9. doi: 10.1074/jbc.M111.264093. Epub 2011 Jul 16. J Biol Chem. 2011. PMID: 21768096 Free PMC article. - Anisotropy in mechanical unfolding of protein upon partner-assisted pulling and handle-assisted pulling.
Arora N, Hazra JP, Rakshit S. Arora N, et al. Commun Biol. 2021 Jul 29;4(1):925. doi: 10.1038/s42003-021-02445-y. Commun Biol. 2021. PMID: 34326473 Free PMC article. - Fluorescence characterization of denatured proteins.
Chen H, Rhoades E. Chen H, et al. Curr Opin Struct Biol. 2008 Aug;18(4):516-24. doi: 10.1016/j.sbi.2008.06.008. Epub 2008 Aug 12. Curr Opin Struct Biol. 2008. PMID: 18675353 Free PMC article. - Thermodynamics of loop formation in the denatured state of rhodopseudomonas palustris cytochrome c': scaling exponents and the reconciliation problem.
Rao KS, Tzul FO, Christian AK, Gordon TN, Bowler BE. Rao KS, et al. J Mol Biol. 2009 Oct 9;392(5):1315-25. doi: 10.1016/j.jmb.2009.07.074. Epub 2009 Aug 6. J Mol Biol. 2009. PMID: 19647747 Free PMC article.
References
- Kuwajima K. Proteins Struct Funct Genet. 1989;6:87–103. - PubMed
- Dobson C M. Curr Opin Struct Biol. 1992;2:6–12.
- Segel D, Bachmann A, Hofrichter J, Hodgson K, Doniach S, Kiefhaber T. J Mol Biol. 1999;288:489–500. - PubMed
- Jackson S E, Fersht A R. Biochemistry. 1991;30:10428–10435. - PubMed
- Alexander P, Orban J, Bryan P. Biochemistry. 1992;31:7243–7248. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous