SMG-2 is a phosphorylated protein required for mRNA surveillance in Caenorhabditis elegans and related to Upf1p of yeast - PubMed (original) (raw)
SMG-2 is a phosphorylated protein required for mRNA surveillance in Caenorhabditis elegans and related to Upf1p of yeast
M F Page et al. Mol Cell Biol. 1999 Sep.
Abstract
mRNAs that contain premature stop codons are selectively degraded in all eukaryotes tested, a phenomenon termed "nonsense-mediated mRNA decay" (NMD) or "mRNA surveillance." NMD may function to eliminate aberrant mRNAs so that they are not translated, because such mRNAs might encode deleterious polypeptide fragments. In both yeasts and nematodes, NMD is a nonessential system. Mutations affecting three yeast UPF genes or seven nematode smg genes eliminate NMD. We report here the molecular analysis of smg-2 of Caenorhabditis elegans. smg-2 is homologous to UPF1 of yeast and to RENT1 (also called HUPF1), a human gene likely involved in NMD. The striking conservation of SMG-2, Upf1p, and RENT1/HUPF1 in both sequence and function suggests that NMD is an ancient system, predating the divergence of most eukaryotes. Despite similarities in the sequences of SMG-2 and Upf1p, expression of Upf1p in C. elegans does not rescue smg-2 mutants. We have prepared anti-SMG-2 polyclonal antibodies and identified SMG-2 on Western blots. SMG-2 is phosphorylated, and mutations of the six other smg genes influence the state of SMG-2 phosphorylation. In smg-1, smg-3, and smg-4 mutants, phosphorylation of SMG-2 was not detected. In smg-5, smg-6, and smg-7 mutants, a phosphorylated isoform of SMG-2 accumulated to abnormally high levels. In smg-2(r866) and smg-2(r895) mutants, which harbor single amino acid substitutions of the SMG-2 nucleotide binding site, phosphorylated SMG-2 accumulated to abnormally high levels, similar to those observed in smg-5, smg-6, and smg-7 mutants. We discuss these results with regard to the in vivo functions of SMG-2 and NMD.
Figures
FIG. 1
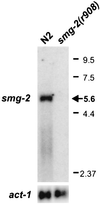
Northern blot hybridized with smg-2 cDNA clone TR#192. smg-2 mRNA is approximately 5.6 kb and is eliminated in smg-2(r908), a deletion that removes all smg-2 sequences. Numbers on the right are molecular sizes, in kilobases.
FIG. 2
smg-2 genomic region. The genomic sequence of the smg-2 region is incomplete, but we deduced a partial restriction map of the region from genomic Southern blots by using either cDNA clone TR#192 or genomic clones TR#178 and TR#179 as hybridization probes. smg-2(r915) is an approximately 1.0-kb deletion. smg-2(r908) and smg-2(r912) delete all sequences contained on cDNA clone TR#192. smg-2(r907) and smg-2(r920) are insertions of Tc1 and Tc4, respectively.
FIG. 3
Sequence of SMG-2. The sequence of SMG-2, deduced from cDNA clone TR#192, is aligned with those of Upf1p and RENT1/HUPF1. Amino acid residues identical in two or three of these three proteins are shaded with light gray and dark gray, respectively. The highly homologous zinc finger and RNA helicase domains are boxed with dotted and solid lines, respectively. The carboxyl termini of SMG-2 and RENT1/HUPF1 are rich in SQ dipeptides, which are underlined.
FIG. 4
Phosphorylation of SMG-2. All panels display total protein Western blots reacted with anti-SMG-2 polyclonal antibodies. (A) Anti-SMG-2 antibodies react with a single protein estimated to be 120 kDa that is absent in smg-2(r908) mutants. (B) Expression of SMG-2 in wild-type animals (N2) and in mutants of all smg genes. All strains were grown at 20°C except the smg-7(r1131) strain, which was grown at both 20 and 25°C because it is temperature sensitive. Parallel experiments demonstrated that the wild type grown at 25°C did not accumulate detectable quantities of the more slowly migrating SMG-2 isoform. (C) Crude extracts (Ext.) of the smg-5(r860) mutant (lane 1) were incubated for 30 min with recombinant lambda protein phosphatase (λPPase) (lane 2) or in phosphatase buffer without added enzyme (lane 3) prior to electrophoresis.
FIG. 5
SMG-2 isoforms of smg smg double mutants. All panels display the SMG-2 region of Western blots reacted with anti-SMG-2 antibody. SMG-2 isoforms which accumulated in smg-5, smg-6, and smg-7 single mutants are shown in row 1. SMG-2 isoforms which accumulated in smg smg double mutants are shown at the intersections of rows and columns. For example, row 2, column 1, is a smg-1 smg-5 double mutant; row 3, column 1, is a smg-3 smg-5 double mutant, etc. Alleles used to construct the double mutants were smg-1(r861), smg-3(r867), smg-4(ma116), smg-5(r860), smg-6(r896), and smg-7(r1131). All strains were grown at 20°C except those containing smg-7(r1131). Because r1131 is temperature sensitive, all strains involving this mutation were grown at 25°C. Experiments not shown demonstrate that growth at 25°C does not influence the relative proportions of SMG-2 and phosphorylated SMG-2 in the wild type or smg-5 and smg-6 single mutants.
FIG. 6
SMG-2 isoforms of smg-2 missense mutations. The SMG-2 region of a Western blot reacted with anti-SMG-2 antibody is shown. Phosphorylated SMG-2 was not detected in the wild type (lane 1) or in two mutants with smg-2 missense alleles (lanes 6 and 7) but was readily detected in smg-2(r866), smg-2(r895), and smg-5 (r860) (control) mutants. smg-2(r908) fully deletes smg-2. Amino acid substitutions predicted from the sequences of smg-2(r866) and smg-2(r895) are shown relative to the consensus sequence of the P-loop of mononucleotide binding proteins (14).
Similar articles
- SMG-5, required for C.elegans nonsense-mediated mRNA decay, associates with SMG-2 and protein phosphatase 2A.
Anders KR, Grimson A, Anderson P. Anders KR, et al. EMBO J. 2003 Feb 3;22(3):641-50. doi: 10.1093/emboj/cdg056. EMBO J. 2003. PMID: 12554664 Free PMC article. - smg-7 is required for mRNA surveillance in Caenorhabditis elegans.
Cali BM, Kuchma SL, Latham J, Anderson P. Cali BM, et al. Genetics. 1999 Feb;151(2):605-16. doi: 10.1093/genetics/151.2.605. Genetics. 1999. PMID: 9927455 Free PMC article. - Caenorhabditis elegans SMG-2 selectively marks mRNAs containing premature translation termination codons.
Johns L, Grimson A, Kuchma SL, Newman CL, Anderson P. Johns L, et al. Mol Cell Biol. 2007 Aug;27(16):5630-8. doi: 10.1128/MCB.00410-07. Epub 2007 Jun 11. Mol Cell Biol. 2007. PMID: 17562857 Free PMC article. - Stop making nonSense: the C. elegans smg genes.
Mango SE. Mango SE. Trends Genet. 2001 Nov;17(11):646-53. doi: 10.1016/s0168-9525(01)02479-9. Trends Genet. 2001. PMID: 11672865 Review. - Role of SMG-1-mediated Upf1 phosphorylation in mammalian nonsense-mediated mRNA decay.
Yamashita A. Yamashita A. Genes Cells. 2013 Mar;18(3):161-75. doi: 10.1111/gtc.12033. Epub 2013 Jan 28. Genes Cells. 2013. PMID: 23356578 Review.
Cited by
- Translation-dependent displacement of UPF1 from coding sequences causes its enrichment in 3' UTRs.
Zünd D, Gruber AR, Zavolan M, Mühlemann O. Zünd D, et al. Nat Struct Mol Biol. 2013 Aug;20(8):936-43. doi: 10.1038/nsmb.2635. Epub 2013 Jul 7. Nat Struct Mol Biol. 2013. PMID: 23832275 - Physiological IRE-1-XBP-1 and PEK-1 signaling in Caenorhabditis elegans larval development and immunity.
Richardson CE, Kinkel S, Kim DH. Richardson CE, et al. PLoS Genet. 2011 Nov;7(11):e1002391. doi: 10.1371/journal.pgen.1002391. Epub 2011 Nov 17. PLoS Genet. 2011. PMID: 22125500 Free PMC article. - SMG-1 is a phosphatidylinositol kinase-related protein kinase required for nonsense-mediated mRNA Decay in Caenorhabditis elegans.
Grimson A, O'Connor S, Newman CL, Anderson P. Grimson A, et al. Mol Cell Biol. 2004 Sep;24(17):7483-90. doi: 10.1128/MCB.24.17.7483-7490.2004. Mol Cell Biol. 2004. PMID: 15314158 Free PMC article. - trt-1 is the Caenorhabditis elegans catalytic subunit of telomerase.
Meier B, Clejan I, Liu Y, Lowden M, Gartner A, Hodgkin J, Ahmed S. Meier B, et al. PLoS Genet. 2006 Feb;2(2):e18. doi: 10.1371/journal.pgen.0020018. Epub 2006 Feb 10. PLoS Genet. 2006. PMID: 16477310 Free PMC article. - Nonsense-mediated mRNA decay factors act in concert to regulate common mRNA targets.
Rehwinkel J, Letunic I, Raes J, Bork P, Izaurralde E. Rehwinkel J, et al. RNA. 2005 Oct;11(10):1530-44. doi: 10.1261/rna.2160905. RNA. 2005. PMID: 16199763 Free PMC article.
References
- Altamura N, Groudinsky O, Dujardin G, Slonimski P P. NAM7 nuclear gene encodes a novel member of a family of helicases with a Zn-ligand motif and is involved in mitochondrial functions in Saccharomyces cerevisiae. J Mol Biol. 1992;224:575–587. - PubMed
- Anders, K., and P. Anderson. Unpublished observations.
- Anderson C W, Lees-Miller S P. The nuclear serine/threonine protein kinase DNA-PK. Crit Rev Eukaryot Gene Expression. 1992;2:283–314. - PubMed
- Anderson P. Mutagenesis. Methods Cell Biol. 1995;48:31–58. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases