Analysis of primary structural determinants that distinguish the centromere-specific function of histone variant Cse4p from histone H3 - PubMed (original) (raw)
Comparative Study
Analysis of primary structural determinants that distinguish the centromere-specific function of histone variant Cse4p from histone H3
K C Keith et al. Mol Cell Biol. 1999 Sep.
Abstract
Cse4p is a variant of histone H3 that has an essential role in chromosome segregation and centromere chromatin structure in budding yeast. Cse4p has a unique 135-amino-acid N terminus and a C-terminal histone-fold domain that is more than 60% identical to histone H3 and the mammalian centromere protein CENP-A. Cse4p and CENP-A have biochemical properties similar to H3 and probably replace H3 in centromere-specific nucleosomes in yeasts and mammals, respectively. In order to identify regions of Cse4p that distinguish it from H3 and confer centromere function, a systematic site-directed mutational analysis was performed. Nested deletions of the Cse4p N terminus showed that this region of the protein contains at least one essential domain. The C-terminal histone-fold domain of Cse4p was analyzed by changing Cse4p amino acids that differ between Cse4p and H3 to the analogous H3 residues. Extensive substitution of contiguous Cse4p residues with H3 counterparts resulted in cell lethality. However, all large lethal substitution alleles could be subdivided into smaller viable alleles, many of which caused elevated rates of mitotic chromosome loss. The results indicate that residues critical for wild-type Cse4p function and high-fidelity chromosome transmission are distributed across the entire histone-fold domain. Our findings are discussed in the context of the known structure of H3 within the nucleosome and compared with previous results reported for CENP-A.
Figures
FIG. 1
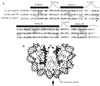
(A) Alignment of the histone-fold domains of S. cerevisiae H3, Cse4p, and human CENP-A. Residues that are identical in two or three of the proteins are indicated in boldface italic type. The α-helices and β-loops in the histone-fold domain of H3 are shown above the residues involved in those structures. The amino acids in H3 involved in dimer formation with H4 are boxed, and the residues involved in the H3-H3′ four-helix bundle interaction are circled. The highly conserved phenylalanine in loop I of H3 that is replaced by a tryptophan (W178) in Cse4p and CENP-A is noted (▴). Specific Cse4p amino acids mentioned in the text are numbered. The positions in the protein sequences of H3 and Cse4p corresponding to the restriction sites used for domain exchanges are indicated (Materials and Methods). (B) The (H3-H4)2 tetramer initiates formation of a standard nucleosome by binding DNA (50 bp are shown). The DNA helix is black, the two copies of H4 are in dark gray, and the two copies of H3 are light gray. The H3-H3′ four-helix bundle is circled, and the vertical arrow indicates the dyad axis. Helix I, II, and III of H3 and H4 are numbered 1, 2, and 3, respectively, and the H3 N-helix is marked with an “N.” The figure was generated by using RasMac version 2.6 with X-ray coordinates from the Protein Data Bank, Brookhaven National Laboratory (ID code 1aoi) (11).
FIG. 2
N-terminal Cse4p deletions reveal an essential domain. (A) Amino acid sequence of the Cse4p N terminus showing deletion endpoints. Mutant alleles initiate with methionine, followed by a histidine residue, and continue with the amino acid indicated. 3× HA indicates the location of the inserted triple HA epitope tag. (B) Plasmid shuffle complementation assays for the cse4 deletion mutants (see Materials and Methods). (C) Immunoblot analysis of yeast strains carrying cse4 deletion alleles. Cells carrying the HA-tagged wild-type allele (WT) were loaded at 1× and 0.5× amounts (all other lanes are 1×) to allow an estimation of signal strength. Lane Vec was loaded with cells carrying the expression vector lacking a CSE4 insert.
FIG. 3
Functional analysis of cse4 alleles with mutations in loop I. The loop I region of Cse4p is aligned with H3 and CENP-A at the bottom of the figure to show the KDQ insert and the position of W178. Mutant cse4 alleles were tested for their ability to complement a cse4 null mutation by using the plasmid shuffle assay (see Materials and Methods). Alleles exhibiting a temperature-sensitive growth phenotype (38°C) are noted (TS).
FIG. 4
Cse4p histone-fold domain mutants. Regions of the histone-fold domain of Cse4p were substituted with the analogous residues from H3. A sequence alignment of the S. cerevisiae H3 and Cse4p proteins is shown at the top of the figure, with identical residues shaded. The positions of the α-helices and β-loops are noted above the residues. Underlined amino acids and asterisks denote regions in H3 that contact DNA in the nucleosome crystal structure. For each mutant allele, the black boxes represent the regions containing substituted H3 residues. The altered Cse4p amino acids are shown above the helical and loop structures, and the substituted H3 amino acids expressed in the mutant are shown below. The viability of the cse4 mutant alleles was determined by using the plasmid shuffle assay. SC, cse4 alleles that reproducibly produce small FOA-resistant colonies in the plasmid shuffle assay (see Materials and Methods).
FIG. 5
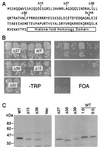
Functional complementation by cse4 histone-fold domain swap mutations. The plasmid shuffle assays (TRP− and FOA plates are as indicated) were performed on cells transformed by the cse4 allele indicated on the right. Asterisks indicate epitope-tagged alleles, WT denotes the wild-type CSE4 allele, and Vec denotes a vector lacking a CSE4 gene. Each of the four rows shows single plates from three independent assays (except for cse4-363, which was taken from a separate plate). The top two sets of plates comprised a single assay, while the bottom sets of plates were separate, independent assays. Growth on Trp− but no growth on FOA indicates lethal cse4 mutations that fail to complement the cse4 null, while growth on FOA plates indicates viable cse4 alleles that can complement the cse4 null. Results from the shuffle assays are summarized in Fig. 4. Several alleles produce small colonies on the FOA plates, a finding indicative of a slower growth rate. These are denoted “SC” in Fig. 4.
FIG. 6
Cse4p histone-fold domain mutants are defective in chromosome segregation. Viable cse4 histone-fold domain mutants were assayed for mitotic loss of chromosome III. Loss rates were determined by fluctuation assays as described in Materials and Methods and are reported as fold increases relative to the wild-type CSE4 control strain.
FIG. 7
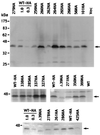
Detection of HA-tagged Cse4 mutant proteins by immunoblotting. Representative blots for viable and lethal cse4 alleles are shown, including alleles used in the mitotic chromosome loss assays. The upper panel shows an entire blot, while the lower panels show only the Cse4p region. The various mutants are identified by the allele numbers above the gel lanes, with HA designating HA-tagged derivatives. Samples of HA-tagged wild-type (WT) cells were loaded at 1× and 0.3× amounts (all other lanes, 1×) to allow an estimation of signal strength. The lane labeled Vec was loaded with cells carrying the expression vector lacking a CSE4 insert. The mobility of protein size markers is indicated at the left, and arrows mark the position of the Cse4p band.
Similar articles
- The N terminus of the centromere H3-like protein Cse4p performs an essential function distinct from that of the histone fold domain.
Chen Y, Baker RE, Keith KC, Harris K, Stoler S, Fitzgerald-Hayes M. Chen Y, et al. Mol Cell Biol. 2000 Sep;20(18):7037-48. doi: 10.1128/MCB.20.18.7037-7048.2000. Mol Cell Biol. 2000. PMID: 10958698 Free PMC article. - Histone-histone interactions and centromere function.
Glowczewski L, Yang P, Kalashnikova T, Santisteban MS, Smith MM. Glowczewski L, et al. Mol Cell Biol. 2000 Aug;20(15):5700-11. doi: 10.1128/MCB.20.15.5700-5711.2000. Mol Cell Biol. 2000. PMID: 10891506 Free PMC article. - The Elusive Structure of Centro-Chromatin: Molecular Order or Dynamic Heterogenetity?
Nagpal H, Fierz B. Nagpal H, et al. J Mol Biol. 2021 Mar 19;433(6):166676. doi: 10.1016/j.jmb.2020.10.010. Epub 2020 Oct 14. J Mol Biol. 2021. PMID: 33065112 Review. - Is there a unique form of chromatin at the Saccharomyces cerevisiae centromeres?
Basrai MA, Hieter P. Basrai MA, et al. Bioessays. 1995 Aug;17(8):669-72. doi: 10.1002/bies.950170802. Bioessays. 1995. PMID: 7661847 Review.
Cited by
- Structure, dynamics, and evolution of centromeric nucleosomes.
Dalal Y, Furuyama T, Vermaak D, Henikoff S. Dalal Y, et al. Proc Natl Acad Sci U S A. 2007 Oct 9;104(41):15974-81. doi: 10.1073/pnas.0707648104. Epub 2007 Sep 24. Proc Natl Acad Sci U S A. 2007. PMID: 17893333 Free PMC article. Review. - Conformational flexibility of histone variant CENP-ACse4 is regulated by histone H4: A mechanism to stabilize soluble Cse4.
Malik N, Dantu SC, Shukla S, Kombrabail M, Ghosh SK, Krishnamoorthy G, Kumar A. Malik N, et al. J Biol Chem. 2018 Dec 28;293(52):20273-20284. doi: 10.1074/jbc.RA118.004141. Epub 2018 Oct 31. J Biol Chem. 2018. PMID: 30381395 Free PMC article. - Scm3, an essential Saccharomyces cerevisiae centromere protein required for G2/M progression and Cse4 localization.
Stoler S, Rogers K, Weitze S, Morey L, Fitzgerald-Hayes M, Baker RE. Stoler S, et al. Proc Natl Acad Sci U S A. 2007 Jun 19;104(25):10571-6. doi: 10.1073/pnas.0703178104. Epub 2007 Jun 4. Proc Natl Acad Sci U S A. 2007. PMID: 17548816 Free PMC article. - Human centromere protein A (CENP-A) can replace histone H3 in nucleosome reconstitution in vitro.
Yoda K, Ando S, Morishita S, Houmura K, Hashimoto K, Takeyasu K, Okazaki T. Yoda K, et al. Proc Natl Acad Sci U S A. 2000 Jun 20;97(13):7266-71. doi: 10.1073/pnas.130189697. Proc Natl Acad Sci U S A. 2000. PMID: 10840064 Free PMC article. - Centromere Structure and Function.
Bloom K, Costanzo V. Bloom K, et al. Prog Mol Subcell Biol. 2017;56:515-539. doi: 10.1007/978-3-319-58592-5_21. Prog Mol Subcell Biol. 2017. PMID: 28840251 Free PMC article. Review.
References
- Bloom K S, Carbon J. Yeast centromere DNA is in a unique and highly ordered structure in chromosomes and small circular minichromosomes. Cell. 1982;29:305–317. - PubMed
- Fitzgerald-Hayes M, Clarke L, Carbon J. Nucleotide sequence comparisons and functional analysis of yeast centromere DNAs. Cell. 1982;29:235–244. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases