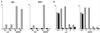Methylation-mediated transcriptional silencing in euchromatin by methyl-CpG binding protein MBD1 isoforms - PubMed (original) (raw)
Methylation-mediated transcriptional silencing in euchromatin by methyl-CpG binding protein MBD1 isoforms
N Fujita et al. Mol Cell Biol. 1999 Sep.
Abstract
DNA methylation of promoter-associated CpG islands is involved in the transcriptional repression of vertebrate genes. To investigate the mechanisms underlying gene inactivation by DNA methylation, we characterized a human MBD1 protein, one of the components of MeCP1, which possesses a methyl-CpG binding domain (MBD) and cysteine-rich (CXXC) domains. Four novel MBD1 isoforms (MBD1v1, MBD1v2, MBD1v3, and MBD1v4) were identified by the reverse transcription-PCR method. We found that these transcripts were alternatively spliced in the region of CXXC domains and the C terminus. Green fluorescent protein-fused MBD1 was localized to multiple foci on the human genome, mostly in the euchromatin regions, and particularly concentrated in the pericentromeric region of chromosome 1. Both the MBD sequence and genome methylation were required for proper localization of the MBD1 protein. We further investigated whether MBD1 isoforms are responsible for transcriptional repression of human genes. A bacterially expressed MBD1 protein bound preferentially to methylated DNA fragments containing CpG islands from the tumor suppressor genes p16, VHL, and E-cadherin and from an imprinted SNRPN gene. All MBD1 isoforms inhibited promoter activities of these genes via methylation. Interestingly, MBD1 isoforms v1 and v2 containing three CXXC domains also suppressed unmethylated promoter activities in mammalian cells. These effects were further manifested in Drosophila melanogaster cells, which lack genome methylation. Sp1-activated transcription of methylated p16 and SNRPN promoters was inhibited by all of the MBD1 isoforms, whereas the isoforms v1 and v2 reduced Sp1-activated transcription from unmethylated promoters as well. These findings suggested that the MBD1 isoforms have different roles in methylation-mediated transcriptional silencing in euchromatin.
Figures
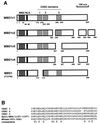
FIG. 1
Protein structure of MBD1 splice isoforms and sequence alignment of the CXXC domains. (A) Diagrams of MBD1 isoforms. These include MBD1v1, MBD1v2, MBD1v3, and MBD1v4 and a previously described MBD1 (formerly PCM1 [Y10746]). They show one methyl-CpG binding domain (MBD), one putative NLS (KKRKK), and two or three cysteine-rich (CXXC) domains, due to alternative splicing events, designated as CXXC1 to CXXC3. The numbers below the diagrams indicate the positions of amino acids (a.a.) from the N terminus. (B) Sequence alignment of the CXXC domains between MBD1 isoforms, ALL1/HRX, and DNA (cytosine-5)-methyltransferase (MTase). Conserved cysteine residues are indicated by boldface type. The accession numbers of the sequences are Q03164 (ALL1/HRX) and X63692 (MTase).
FIG. 2
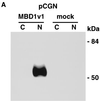
Subcellular localization of MBD1 protein. (A) Western blot analysis. pCGN-MBD1v1 (amino acids 1 to 421) was transfected into COS-7 cells. HA-tagged MBD1 was present in the nuclear fraction (N) but not in the cytosolic fraction (C). (B to F) EGFP-fused MBD1 was transiently expressed in COS-7 and NCI-H1299 cells and observed with a CLSM. (B to E) The lower row shows a transmitted view (Nomarski) of the upper. (B) The result of the full-length MBD1v1 is shown in the upper row. Three vectors were constructed: pEGFP-MBD1(MBD+NLS), expressing an EGFP fused to both MBD and putative NLS (C); pEGFP-MBD1(MBD), expressing an EGFP fused to MBD alone (D); and pEGFP-MBD1(NLS), expressing an EGFP fused to putative NLS alone (E). (F) Localization of EGFP-MBD1(MBD+NLS) in the nuclei of NCI-H1299 cells which were treated with 5-aza-2′-deoxycytidine (0.5 μM) for 6 days. The methylation-specific PCR with bisulfite modification of DNA (lower panels) demonstrated the demethylation of the promoter region of human p16 gene after the treatment. W, U and M indicate specifically amplified fragments corresponding to unmodified, unmethylated, and methylated sequences, respectively.
FIG. 2
Subcellular localization of MBD1 protein. (A) Western blot analysis. pCGN-MBD1v1 (amino acids 1 to 421) was transfected into COS-7 cells. HA-tagged MBD1 was present in the nuclear fraction (N) but not in the cytosolic fraction (C). (B to F) EGFP-fused MBD1 was transiently expressed in COS-7 and NCI-H1299 cells and observed with a CLSM. (B to E) The lower row shows a transmitted view (Nomarski) of the upper. (B) The result of the full-length MBD1v1 is shown in the upper row. Three vectors were constructed: pEGFP-MBD1(MBD+NLS), expressing an EGFP fused to both MBD and putative NLS (C); pEGFP-MBD1(MBD), expressing an EGFP fused to MBD alone (D); and pEGFP-MBD1(NLS), expressing an EGFP fused to putative NLS alone (E). (F) Localization of EGFP-MBD1(MBD+NLS) in the nuclei of NCI-H1299 cells which were treated with 5-aza-2′-deoxycytidine (0.5 μM) for 6 days. The methylation-specific PCR with bisulfite modification of DNA (lower panels) demonstrated the demethylation of the promoter region of human p16 gene after the treatment. W, U and M indicate specifically amplified fragments corresponding to unmodified, unmethylated, and methylated sequences, respectively.
FIG. 2
Subcellular localization of MBD1 protein. (A) Western blot analysis. pCGN-MBD1v1 (amino acids 1 to 421) was transfected into COS-7 cells. HA-tagged MBD1 was present in the nuclear fraction (N) but not in the cytosolic fraction (C). (B to F) EGFP-fused MBD1 was transiently expressed in COS-7 and NCI-H1299 cells and observed with a CLSM. (B to E) The lower row shows a transmitted view (Nomarski) of the upper. (B) The result of the full-length MBD1v1 is shown in the upper row. Three vectors were constructed: pEGFP-MBD1(MBD+NLS), expressing an EGFP fused to both MBD and putative NLS (C); pEGFP-MBD1(MBD), expressing an EGFP fused to MBD alone (D); and pEGFP-MBD1(NLS), expressing an EGFP fused to putative NLS alone (E). (F) Localization of EGFP-MBD1(MBD+NLS) in the nuclei of NCI-H1299 cells which were treated with 5-aza-2′-deoxycytidine (0.5 μM) for 6 days. The methylation-specific PCR with bisulfite modification of DNA (lower panels) demonstrated the demethylation of the promoter region of human p16 gene after the treatment. W, U and M indicate specifically amplified fragments corresponding to unmodified, unmethylated, and methylated sequences, respectively.
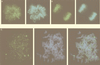
FIG. 3
Intranuclear territories of MBD1 and other chromosomal proteins in human HeLa cells. EGFP-MBD1(MBD+NLS) (green) transfected into HeLa cells produced a punctate staining pattern in the nucleus (A and C), while EGFP-MeCP2 (green) was distributed throughout the nucleus (B). Chromosomal DNAs were counterstained with propidium iodide (red) (A and B). MBD1(MBD+NLS) did not colocalize with kinetochores, which were labeled with autoantibodies from a patient with the CREST syndrome of scleroderma (red) (C). The right-hand panels show merges of the left-hand and the middle panels.
FIG. 4
Localization of MBD1 on human chromosomes. EGFP-MBD1(MBD+NLS) (green) was found in HeLa cells on the metaphase chromosomes (A), on the chromosomes during mitosis (B), and on the metaphase chromosome spreads (C), using a deconvolution system with a highly sensitive charge-coupled camera. Chromosomal DNAs, especially in the heterochromatin regions, were counterstained with DAPI (blue) (A to C). (C) MBD1 localized mostly in negatively DAPI-stained euchromatic regions on human chromosomes and to pericentromeric regions of chromosome 1. The right-hand panel shows a merge of left-hand and middle panels.
FIG. 5
Effect of MBD1 on promoter-associated CpG islands of p16, VHL, E-cadherin, and SNRPN genes. (A) PCR-amplified DNA fragments from the gene promoters were subcloned upstream of a luciferase cDNA in a pGL3-Basic vector. The position of the transcription start site and the oligonucleotide primers DF and ER to amplify DNA fragments utilized for the in vitro transcription assay are shown by solid and open arrows, respectively. The presence of CpG dinucleotides within the inserted promoter sequences of four genes is indicated by vertical lines. (B) Band shift of methylated DNAs complexed with MBD1. Unmethylated (M−) and methylated (M+) DNA fragments containing CpG islands from these genes were incubated with GST-fused MBD1v1 or GST alone. The in vitro methylation of CpG sequences was performed with _Sss_I methyltransferase. +, present; −, absent. (C) Transcriptional repression by MBD1 in a methylation-dependent manner. GST-MBD1 was incubated with either unmethylated (M−) or methylated (M+) DNA fragments which were PCR amplified from the promoter-inserted pGL3 vectors with primers DF and ER. Transcripts from the promoters were synthesized in HeLa nuclear extract and detected by the primer extension method with a radiolabeled ER primer. The amount of a predicted cDNA product was measured by a Bioimaging analyzer, MacBAS version 2.51, and the relative amount of transcript compared with that of the unmethylated promoter without GST-MBD1 is indicated below each panel.
FIG. 6
Transcriptional repression by MBD1 isoforms in mammalian cells. (A) Expression of HA-tagged MBD1 isoforms. Four vectors expressing MBD1 isoforms, pCGN-MBD1v1 to pCGN-MBD1v4, were transfected into CHO cells, and a Western blot analysis with an anti-HA epitope monoclonal antibody was performed. (B) Inhibition of p16, VHL, E-cadherin, and SNRPN gene promoter activities by MBD1 isoforms. Both promoter-inserted pGL3 and pCGN-MBD1 vectors in the appropriate combinations were cotransfected into CHO cells. The level of luciferase activity in the cotransfection of unmethylated promoter-inserted pGL3 and pCGN (mock) was normalized to 10,000 in each gene promoter. The average of relative activities from four independent experiments is indicated by each bar. Unmethylated (M−) and methylated (M+) promoter-inserted pGL3 vectors are shown in the upper and lower rows, respectively.
FIG. 6
Transcriptional repression by MBD1 isoforms in mammalian cells. (A) Expression of HA-tagged MBD1 isoforms. Four vectors expressing MBD1 isoforms, pCGN-MBD1v1 to pCGN-MBD1v4, were transfected into CHO cells, and a Western blot analysis with an anti-HA epitope monoclonal antibody was performed. (B) Inhibition of p16, VHL, E-cadherin, and SNRPN gene promoter activities by MBD1 isoforms. Both promoter-inserted pGL3 and pCGN-MBD1 vectors in the appropriate combinations were cotransfected into CHO cells. The level of luciferase activity in the cotransfection of unmethylated promoter-inserted pGL3 and pCGN (mock) was normalized to 10,000 in each gene promoter. The average of relative activities from four independent experiments is indicated by each bar. Unmethylated (M−) and methylated (M+) promoter-inserted pGL3 vectors are shown in the upper and lower rows, respectively.
FIG. 7
Transcriptional regulation by MBD1 isoforms in Drosophila SL2 cells. (A) Unmethylated (M−) or methylated (M+) pGL3 construct (pGL3-p16 and pGL3-SNRPN or pGL3-VHL and pGL3-SNRPN) was cotransfected in combination with mock A5C vector (solid bars), pPacSp1 expressing transcription factor Sp1 (hatched bars on the left), or pAc5.1-E2F1 expressing transcription factor E2F1 (hatched bars on the right) into SL2 cells. The level of luciferase activity in the presence of unmethylated pGL3 and mock vectors was normalized to 1 for each gene promoter. (B) Methylated (solid bars) or unmethylated (hatched bars) promoter-inserted pGL3 vector was cotransfected with pPacSp1 and MBD1-expressing plasmids (pAc5.1-MBD1v1 to pAc5.1-MBD1v4) or insertless plasmid (mock). The luciferase activity of unmethylated pGL3 in the combination of pPacSp1 and mock was normalized to 100 for each gene promoter, and the relative luciferase activities are presented.
Similar articles
- Mechanism of transcriptional regulation by methyl-CpG binding protein MBD1.
Fujita N, Shimotake N, Ohki I, Chiba T, Saya H, Shirakawa M, Nakao M. Fujita N, et al. Mol Cell Biol. 2000 Jul;20(14):5107-18. doi: 10.1128/MCB.20.14.5107-5118.2000. Mol Cell Biol. 2000. PMID: 10866667 Free PMC article. - Regulation of transcription and chromatin by methyl-CpG binding protein MBD1.
Nakao M, Matsui S, Yamamoto S, Okumura K, Shirakawa M, Fujita N. Nakao M, et al. Brain Dev. 2001 Dec;23 Suppl 1:S174-6. doi: 10.1016/s0387-7604(01)00348-5. Brain Dev. 2001. PMID: 11738867 Review. - Mbd1 is recruited to both methylated and nonmethylated CpGs via distinct DNA binding domains.
Jørgensen HF, Ben-Porath I, Bird AP. Jørgensen HF, et al. Mol Cell Biol. 2004 Apr;24(8):3387-95. doi: 10.1128/MCB.24.8.3387-3395.2004. Mol Cell Biol. 2004. PMID: 15060159 Free PMC article. - Methyl-CpG binding domain proteins and their involvement in the regulation of the MAGE-A1, MAGE-A2, MAGE-A3, and MAGE-A12 gene promoters.
Wischnewski F, Friese O, Pantel K, Schwarzenbach H. Wischnewski F, et al. Mol Cancer Res. 2007 Jul;5(7):749-59. doi: 10.1158/1541-7786.MCR-06-0364. Mol Cancer Res. 2007. PMID: 17634428 - Methyl-CpG-binding proteins. Targeting specific gene repression.
Ballestar E, Wolffe AP. Ballestar E, et al. Eur J Biochem. 2001 Jan;268(1):1-6. doi: 10.1046/j.1432-1327.2001.01869.x. Eur J Biochem. 2001. PMID: 11121095 Review.
Cited by
- Epigenetic Aberrations in Major Psychiatric Diseases Related to Diet and Gut Microbiome Alterations.
Nohesara S, Abdolmaleky HM, Thiagalingam S. Nohesara S, et al. Genes (Basel). 2023 Jul 24;14(7):1506. doi: 10.3390/genes14071506. Genes (Basel). 2023. PMID: 37510410 Free PMC article. Review. - Inhibitors of DNA Methylation.
Lopez M, Gilbert J, Contreras J, Halby L, Arimondo PB. Lopez M, et al. Adv Exp Med Biol. 2022;1389:471-513. doi: 10.1007/978-3-031-11454-0_17. Adv Exp Med Biol. 2022. PMID: 36350520 Review. - Proteins That Read DNA Methylation.
Liu K, Shimbo T, Song X, Wade PA, Min J. Liu K, et al. Adv Exp Med Biol. 2022;1389:269-293. doi: 10.1007/978-3-031-11454-0_11. Adv Exp Med Biol. 2022. PMID: 36350514 - Optochemical Control of TET Dioxygenases Enables Kinetic Insights into the Domain-Dependent Interplay of TET1 and MBD1 while Oxidizing and Reading 5-Methylcytosine.
Lin TC, Palei S, Summerer D. Lin TC, et al. ACS Chem Biol. 2022 Jul 15;17(7):1844-1852. doi: 10.1021/acschembio.2c00245. Epub 2022 Jun 16. ACS Chem Biol. 2022. PMID: 35709470 Free PMC article. - In silico prediction and interaction of resveratrol on methyl-CpG binding proteins by molecular docking and MD simulations study.
Sahu RK, Verma VV, Kumar A, Tandon S, Chandra Das B, Hedau ST. Sahu RK, et al. RSC Adv. 2022 Apr 13;12(18):11493-11504. doi: 10.1039/d2ra00432a. eCollection 2022 Apr 7. RSC Adv. 2022. PMID: 35425086 Free PMC article.
References
- Ballabio A, Willard H F. Mammalian X-chromosome inactivation and the XIST gene. Curr Opin Genet Dev. 1992;2:439–447. - PubMed
- Bartolomei M S, Tilghman S M. Genomic imprinting in mammals. Annu Rev Genet. 1997;31:493–525. - PubMed
- Baylin S B, Herman J G, Graff J R, Vertino P M, Issa J P. Alterations in DNA methylation: a fundamental aspect of neoplasia. Adv Cancer Res. 1998;72:141–196. - PubMed
- Bergman Y, Mostoslavsky R. DNA demethylation: turning genes on. Biol Chem. 1998;379:401–407. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials