Oxidative damage to the c-fos gene and reduction of its transcription after focal cerebral ischemia - PubMed (original) (raw)
Oxidative damage to the c-fos gene and reduction of its transcription after focal cerebral ischemia
J Cui et al. J Neurochem. 1999 Sep.
Abstract
We investigated oxidative damage to the c-fos gene and to its transcription in the brain of Long-Evans rats using a transient focal cerebral ischemia and reperfusion (FCIR) model. We observed a significant (p < 0.001) increase in the immunoreactivity to 8-hydroxy-2'-guanine (oh8G) and its deoxy form (oh8dG) in the ischemic cortex at 0-30 min of reperfusion in all 27 animals treated with 15-90 min of ischemia. Treatment with a neuronal nitric oxide synthase (nNOS) inhibitor, 3-bromo-7-nitroindazole (60 mg/kg, i.p.), abolished the majority but not all of the oh8G/oh8dG immunoreactivity. Treatment with RNase A reduced the oh8G immunoreactivity, suggesting that RNA may be targeted. This observation was further supported by decreased levels of mRNA transcripts of the c-fos and actin genes in the ischemic core within 30 min of reperfusion using in situ hybridization. The reduction in mRNA transcription occurred at a time when nuclear gene damage, detected as sensitive sites to Escherichia coli Fpg protein in the transcribed strand of the c-fos gene, was increased 13-fold (p < 0.01). Our results suggest that inhibiting nNOS partially attenuates FCIR-induced oxidative damage and that nNOS or other mechanisms induce nuclear gene damage that interferes with gene transcription in the brain.
Figures
FIG. 1
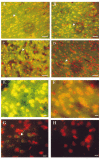
oh8G/oh8dG immunoreactivity after RNase A or 3BR7NI. The fluorescent signal of oh8G/oh8dG in the right (ischemic or ipsilateral) cortices of six animals following FCIR (A—G) and one of the non-FCI (control) animals (H) is shown. Nuclei were counterstained using PI in the presence of RNase A (20 _μ_g/ml, 5 min at room temperature) and appear as red fluorescent signals. The yellowish fluorescent signals indicate colocalization of the oh8dG immunoreactivity with the nuclear PI staining. Two typical cortical samples with 15/0 (A) and 60/0 (B) without reperfusion are shown, along with 90 min of FCI and 15 min of reperfusion (90/15 FCIR in C and D), except the brain samples in D were further treated with RNase A (5 mg/ml,1hat 37°C). A higher magnification is shown of the right cortical sample from one of three animals with 90/15 FCIR and oil (E) or 3BR7NI (F). Another FCIR sample (90/30) is also shown in G. Bar = 30 _μ_m in A—D and 10 _μ_m in E—H.
FIG. 2
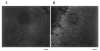
Elevation of oh8G/oh8dG immunoreactivity in the cortex. The left (A; contralateral) and right (B; ischemic) cerebral cortices from one of six animals treated with 90/15 FCIR are shown in black and white. The oh8dG immunoreactivity appears as a white fluorescent signal. Bar = 120 _μ_m.
FIG. 3
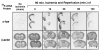
Expression of the c-fos and actin genes using ISH. The expression of the c-fos and actin genes in the cerebral cortex was determined for six treatment groups in a total of 23 animals. Radiograms of four coronal sections per end point (two examined for c-fos and two examined for actin, rostral-to-caudal view; sections in each box are 0.2 mm apart) from one representative animal in each group are shown. Each of the coronal sections used for the c-fos transcript detection was adjacent to the respective section used for the actin transcript detection.
FIG. 4
Quantification of c-fos and actin mRNA transcripts. The transcript intensity in the brain sections from the animals at each time point by ISH was measured using an image documentation system (AlphaImager; Alpha Innotech, San Leandro, CA, U.S.A.). At least two brain sections from each animal were examined in repeated ISH. Data are mean ± SEM (bars) values; the number of animals (n) was n = 3 each for control, 90/0, and 90/30 FCIR, n = 6 for 90/15 FCIR, and n = 2 for each for 90/60 and 90/120 FCIR. Figure 3 shows the typical expression of these two genes at each time point.
FIG. 5
Presence of FPGSS in the c-fos gene. FPGSS were measured in genomic DNA from right cortices of nine sham-operated control animals and from right (ischemic) cortices of 20 rats that underwent 90 min of FCI and various durations of reperfusion. The Fpg protein-treated DNA samples (Y) were resolved in parallel with DNA samples with no Fpg protein treatment (N). The blots were hybridized to 32P-mRNA (for the transcribed strand) or 32P-cRNA (for the nontranscribed strand) of the c-fos gene.
FIG. 6
FPGSS frequencies in the c-fos gene. The signal intensity in the autoradiogram was measured using the AlphaImager. The pixel value of the autoradiogram background was subtracted. The genomic FPGSS frequency was calculated as described in Materials and Methods. The frequencies of genomic FPGSS in the c-fos gene at 15 and 30 min after 90 min of FCI were significantly different (F = 27) using two-way ANOVA: *p < 0.001.
Similar articles
- Oxidative DNA damage precedes DNA fragmentation after experimental stroke in rat brain.
Cui J, Holmes EH, Greene TG, Liu PK. Cui J, et al. FASEB J. 2000 May;14(7):955-67. doi: 10.1096/fasebj.14.7.955. FASEB J. 2000. PMID: 10783150 Free PMC article. - Expression of c-fos and c-jun family genes after focal cerebral ischemia.
An G, Lin TN, Liu JS, Xue JJ, He YY, Hsu CY. An G, et al. Ann Neurol. 1993 May;33(5):457-64. doi: 10.1002/ana.410330508. Ann Neurol. 1993. PMID: 7684582 - Brain injury by global ischemia and reperfusion: a theoretical perspective on membrane damage and repair.
White BC, Grossman LI, Krause GS. White BC, et al. Neurology. 1993 Sep;43(9):1656-65. doi: 10.1212/wnl.43.9.1656. Neurology. 1993. PMID: 8414008 Review. No abstract available.
Cited by
- Noninvasive detection of neural progenitor cells in living brains by MRI.
Liu CH, Ren JQ, You Z, Yang J, Liu CM, Uppal R, Liu PK. Liu CH, et al. FASEB J. 2012 Apr;26(4):1652-62. doi: 10.1096/fj.11-199547. Epub 2011 Dec 23. FASEB J. 2012. PMID: 22198388 Free PMC article. - DNA methyltransferase contributes to delayed ischemic brain injury.
Endres M, Meisel A, Biniszkiewicz D, Namura S, Prass K, Ruscher K, Lipski A, Jaenisch R, Moskowitz MA, Dirnagl U. Endres M, et al. J Neurosci. 2000 May 1;20(9):3175-81. doi: 10.1523/JNEUROSCI.20-09-03175.2000. J Neurosci. 2000. PMID: 10777781 Free PMC article. - Ischemic tolerance as an active and intrinsic neuroprotective mechanism.
Stetler RA, Zhang F, Liu C, Chen J. Stetler RA, et al. Handb Clin Neurol. 2009;92:171-95. doi: 10.1016/S0072-9752(08)01909-X. Handb Clin Neurol. 2009. PMID: 18790275 Free PMC article. Review. No abstract available. - Chronic oxidative damage together with genome repair deficiency in the neurons is a double whammy for neurodegeneration: Is damage response signaling a potential therapeutic target?
Wang H, Dharmalingam P, Vasquez V, Mitra J, Boldogh I, Rao KS, Kent TA, Mitra S, Hegde ML. Wang H, et al. Mech Ageing Dev. 2017 Jan;161(Pt A):163-176. doi: 10.1016/j.mad.2016.09.005. Epub 2016 Sep 20. Mech Ageing Dev. 2017. PMID: 27663141 Free PMC article. Review. - Homogeneous repair of nuclear genes after experimental stroke.
Moore N, Okocha F, Cui JK, Liu PK. Moore N, et al. J Neurochem. 2002 Jan;80(1):111-8. doi: 10.1046/j.0022-3042.2001.00680.x. J Neurochem. 2002. PMID: 11796749 Free PMC article.
References
- An G, Lin T, Liu JS, Xue JJ, He YY, Hsu CY. Expression of c-fos and c-jun family genes after focal cerebral ischemia. Ann. Neurol. 1993;33:457–464. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical