The 17-gene ethanolamine (eut) operon of Salmonella typhimurium encodes five homologues of carboxysome shell proteins - PubMed (original) (raw)
The 17-gene ethanolamine (eut) operon of Salmonella typhimurium encodes five homologues of carboxysome shell proteins
E Kofoid et al. J Bacteriol. 1999 Sep.
Abstract
The eut operon of Salmonella typhimurium encodes proteins involved in the cobalamin-dependent degradation of ethanolamine. Previous genetic analysis revealed six eut genes that are needed for aerobic use of ethanolamine; one (eutR), encodes a positive regulator which mediates induction of the operon by vitamin B12 plus ethanolamine. The DNA sequence of the eut operon included 17 genes, suggesting a more complex pathway than that revealed genetically. We have correlated an open reading frame in the sequence with each of the previously identified genes. Nonpolar insertion and deletion mutations made with the Tn10-derived transposable element T-POP showed that at least 10 of the 11 previously undetected eut genes have no Eut phenotype under the conditions tested. Of the dispensable eut genes, five encode apparent homologues of proteins that serve (in other organisms) as shell proteins of the carboxysome. This bacterial organelle, found in photosynthetic and sulfur-oxidizing bacteria, may contribute to CO2 fixation by concentrating CO2 and excluding oxygen. The presence of these homologues in the eut operon of Salmonella suggests that CO2 fixation may be a feature of ethanolamine catabolism in Salmonella.
Figures
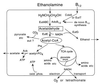
FIG. 1
Suggested pathway for metabolism of ethanolamine. Gene assignments are based on direct assays (EutBCE and CobA), on mutant phenotypes (EutA, EutT, Ack, Pta, and glyoxylate shunt), or on sequence similarity (EutG). The homologues of carboxysome shell proteins suggest the possibility of CO2 fixation, which has not been demonstrated. In the diagram, the outer boundary is the cell membrane; the role of carboxysomes in this pathway is unknown.
FIG. 2
Diagram of the eut operon sequence. The genes underlined in black were discovered by genetic characterization of mutants defective for aerobic use of ethanolamine. The regions underlined in gray were sequenced previously (27, 60). The transposon shown in the E. coli sequence was found in one isolate of E. coli (8) but not in another (69).
FIG. 3
Alignment of carboxysome shell protein homologues. The top panel shows the alignment of the EutN protein with the CcmL protein of Synechococcus. The middle panel aligns the EutK and EutM proteins with the CcmK protein of Synechococcus and the PduA protein of the Salmonella pdu (propanediol utilization) operon. The bottom panel aligns the EutL and EutS proteins with the PduB protein from the Salmonella pdu operon. The EutLS family is most similar to the PduA protein of the EutK, EutN, CcmK class. The sequence features shared by the EutLS-PduB family and the PduA protein are indicated by the black dots below the sequences.
FIG. 4
Sequence motif that EutJ and EutA proteins share with the chaperonin DnaK. Only EutJ shows significant similarity to DnaK over its entire length; EutJ and EutA are not significantly similar to each other, but they share the motif mentioned above. The central DIGGT motif is part of a nucleotide binding loop in the DnaK protein (55).
Similar articles
- Functions required for vitamin B12-dependent ethanolamine utilization in Salmonella typhimurium.
Roof DM, Roth JR. Roof DM, et al. J Bacteriol. 1989 Jun;171(6):3316-23. doi: 10.1128/jb.171.6.3316-3323.1989. J Bacteriol. 1989. PMID: 2656649 Free PMC article. - Autogenous regulation of ethanolamine utilization by a transcriptional activator of the eut operon in Salmonella typhimurium.
Roof DM, Roth JR. Roof DM, et al. J Bacteriol. 1992 Oct;174(20):6634-43. doi: 10.1128/jb.174.20.6634-6643.1992. J Bacteriol. 1992. PMID: 1328159 Free PMC article. - FNR and its role in oxygen-regulated gene expression in Escherichia coli.
Spiro S, Guest JR. Spiro S, et al. FEMS Microbiol Rev. 1990 Aug;6(4):399-428. doi: 10.1111/j.1574-6968.1990.tb04109.x. FEMS Microbiol Rev. 1990. PMID: 2248796 Review. - Prokaryotic Organelles: Bacterial Microcompartments in E. coli and Salmonella.
Stewart KL, Stewart AM, Bobik TA. Stewart KL, et al. EcoSal Plus. 2020 Oct;9(1):10.1128/ecosalplus.ESP-0025-2019. doi: 10.1128/ecosalplus.ESP-0025-2019. EcoSal Plus. 2020. PMID: 33030141 Free PMC article. Review.
Cited by
- Using comparative genomics to uncover new kinds of protein-based metabolic organelles in bacteria.
Jorda J, Lopez D, Wheatley NM, Yeates TO. Jorda J, et al. Protein Sci. 2013 Feb;22(2):179-95. doi: 10.1002/pro.2196. Epub 2013 Jan 4. Protein Sci. 2013. PMID: 23188745 Free PMC article. - Microcompartments in prokaryotes: carboxysomes and related polyhedra.
Cannon GC, Bradburne CE, Aldrich HC, Baker SH, Heinhorst S, Shively JM. Cannon GC, et al. Appl Environ Microbiol. 2001 Dec;67(12):5351-61. doi: 10.1128/AEM.67.12.5351-5361.2001. Appl Environ Microbiol. 2001. PMID: 11722879 Free PMC article. Review. No abstract available. - Short N-terminal sequences package proteins into bacterial microcompartments.
Fan C, Cheng S, Liu Y, Escobar CM, Crowley CS, Jefferson RE, Yeates TO, Bobik TA. Fan C, et al. Proc Natl Acad Sci U S A. 2010 Apr 20;107(16):7509-14. doi: 10.1073/pnas.0913199107. Epub 2010 Mar 22. Proc Natl Acad Sci U S A. 2010. PMID: 20308536 Free PMC article. - Deciphering molecular details in the assembly of alpha-type carboxysome.
Liu Y, He X, Lim W, Mueller J, Lawrie J, Kramer L, Guo J, Niu W. Liu Y, et al. Sci Rep. 2018 Oct 10;8(1):15062. doi: 10.1038/s41598-018-33074-x. Sci Rep. 2018. PMID: 30305640 Free PMC article. - Genome sequence and analysis of the oral bacterium Fusobacterium nucleatum strain ATCC 25586.
Kapatral V, Anderson I, Ivanova N, Reznik G, Los T, Lykidis A, Bhattacharyya A, Bartman A, Gardner W, Grechkin G, Zhu L, Vasieva O, Chu L, Kogan Y, Chaga O, Goltsman E, Bernal A, Larsen N, D'Souza M, Walunas T, Pusch G, Haselkorn R, Fonstein M, Kyrpides N, Overbeek R. Kapatral V, et al. J Bacteriol. 2002 Apr;184(7):2005-18. doi: 10.1128/JB.184.7.2005-2018.2002. J Bacteriol. 2002. PMID: 11889109 Free PMC article.
References
- Babior B M, Carty T J, Abeles R H. The mechanism of action of ethanolamine ammonia-lyase, a B12-dependent enzyme. J Biol Chem. 1974;249:1689–1695. - PubMed
- Baker S H, Jin S, Aldrich H C, Howard G T, Shively J M. Insertion mutation of the form I cbbL gene encoding ribulose bisphosphate carboxylase/oxygenase (RuBisCO) in Thiobacillus neapolitanus results in expression of form II RuBisCO, loss of carboxysomes, and an increased CO2 requirement for growth. J Bacteriol. 1998;180:4133–4139. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases