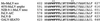The role of the membrane-spanning domain sequence in glycoprotein-mediated membrane fusion - PubMed (original) (raw)
The role of the membrane-spanning domain sequence in glycoprotein-mediated membrane fusion
G M Taylor et al. Mol Biol Cell. 1999 Sep.
Free PMC article
Abstract
The role of glycoprotein membrane-spanning domains in the process of membrane fusion is poorly understood. It has been demonstrated that replacing all or part of the membrane-spanning domain of a viral fusion protein with sequences that encode signals for glycosylphosphatidylinositol linkage attachment abrogates membrane fusion activity. It has been suggested, however, that the actual amino acid sequence of the membrane-spanning domain is not critical for the activity of viral fusion proteins. We have examined the function of Moloney murine leukemia virus envelope proteins with substitutions in the membrane-spanning domain. Envelope proteins bearing substitutions for proline 617 are processed and incorporated into virus particles normally and bind to the viral receptor. However, they possess greatly reduced or undetectable capacities for the promotion of membrane fusion and infectious virus particle formation. Our results imply a direct role for the residues in the membrane-spanning domain of the murine leukemia virus envelope protein in membrane fusion and its regulation. They also support the thesis that membrane-spanning domains possess a sequence-dependent function in other protein-mediated membrane fusion events.
Figures
Figure 1
Amino acid sequence of the ecotropic Mo-MuLV membrane-spanning domain and surrounding residues of TM compared with that of other murine, feline, and gibbon ape leukemia viruses (GeneBank accession numbers 74692, M33469, K02730, 74707, 74702, and M26927, respectively). The absolutely conserved residues in the putative membrane-spanning domain are represented by the bold type, whereas the specific residues that were mutated are indicated by an asterisk.
Figure 2
The mutant envelope proteins are processed normally in NIH 3T3 cells, with the exception of F607V. Cells (5 × 105) that were stably expressing the various mutant envelope proteins were labeled with 50 μCi/ml 35S-cysteine/[35S]methionine for 4 h before lysis with 1× RIP buffer. The cell debris was removed before two rounds of immunoprecipitation of the envelope proteins with antibody against SU. The immunoprecipitated envelope proteins were analyzed on an 8% SDS-PAGE gel and exposed to autoradiography film for 1 wk. Some variation in expression level from cells with different sites of stable env gene integration is the expected result. The amino acid substitutions in the mutant envelope proteins expressed in the lysates are indicated by their representations in the single-letter amino acid code beneath the headings noting the altered residues. GP+E-86 cells were used as the positive control for expression of wild-type Mo-MuLV envelope protein (w.t.), whereas NIH 3T3 cells were used as the negative control (−). At left are indicated the positions of wild-type uncleaved SU+TM (85 kDa) and SU (70 kDa). There is a cross-reactive protein in the cell lysate that migrates slightly slower than SU+TM (85 kDa).
Figure 3
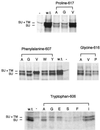
Analysis of the incorporation into virus particles (A) and processing (B) of the various mutant Mo-MuLV envelope proteins in gpGFP cells. gpGFP cells (5 × 105) were transiently transfected with plasmids encoding the mutant and wild-type envelope proteins 48 h before labeling with 35S-cysteine/[35S]methionine for 16 h. The amino acid substitutions in the mutant envelope proteins expressed in the transfected cells are indicated by their representations in the single-letter amino acid code beneath the headings noting the altered residues. Analyses of cell lysates and virus particles budded from cells transfected with the plasmid encoding the wild-type Mo-MuLV envelope protein (w.t.) and from cells that were mock transfected (−) are also presented. At left is indicated the position of SU (70 kDa). (A) Immunoprecipitation of the cell supernatant medium was carried out by filtering it through a 0.45-μm filter and spinning it through a 30% sucrose cushion for 2 h at 25,000 rpm and 4°C. The envelope proteins were immunoprecipitated with antibody against SU, and they were analyzed as above. There is a cross-reactive protein that migrates slightly slower than SU (70 kDa). (B) Immunoprecipitation of the cell lysate was carried out by lysing the cells with 1× RIP buffer. The cell debris was removed before two rounds of immunoprecipitation of the envelope proteins with antibody against SU. The immunoprecipitated envelope proteins were analyzed on an 8% SDS-PAGE gel and exposed to autoradiography film for 1 wk.
Figure 4
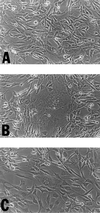
Analysis of the ability of the W606S mutant envelope protein to undergo receptor-mediated-membrane fusion in the presence of the full-length cytoplasmic domain. Four micrograms of plasmids (A) penv1min, (B) penvΔ650–665, and (C) penvW606S were transiently transfected into 5 × 105 NIH 3T3 cells. The transfected cells were allowed to grow for 36 h before syncytia were recorded. Syncytia can be observed in B and C but not in A.
Figure 5
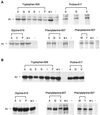
Analysis of the processing of the proline 617 mutants with and without a full-length cytoplasmic domain in 293T cells. 293T cells (1 × 106) were transiently transfected with plasmids encoding the mutant and wild-type envelope proteins with or without the full-length cytoplasmic domain 48 h before labeling with 35S-cysteine/[35S]methionine for 4 h. The amino acid substitutions in the mutant envelope proteins expressed in the transfected cells are indicated by their representations in the single-letter amino acid code beneath the heading noting the altered residue. Analysis of the lysates from cells transfected with the plasmids encoding the wild-type Mo-MuLV envelope protein with the full-length cytoplasmic domain (w.t.) or without the full-length cytoplasmic domain (Δ650–665) and from cells that were mock transfected (−) are also presented. At left is indicated the positions of SU+TM (85 kDa) and SU (70 kDa). Immunoprecipitation and analysis by gel electrophoresis and autoradiography were conducted as described in Figure 3B. The mobility of the SU+TM of proteins with the cytoplasmic domain truncation is increased, whereas that of the mature SU is unaffected.
Figure 6
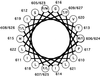
Helical wheel representation of the proposed α-helical membrane-spanning domain of Mo-MuLV. The helical wheel is a depiction of an ideal α-helix with 3.6 residues per turn and a repeating unit of 18 residues or five complete turns. The residues whose role was examined in this article are indicated by the outlined numbers and letters. Glycine 616 is on the polar face of the helix, whereas proline 617 is at the interface between the polar face and apolar face.
Similar articles
- Membrane-proximal cytoplasmic domain of Moloney murine leukemia virus envelope tail facilitates fusion.
Rozenberg-Adler Y, Conner J, Aguilar-Carreno H, Chakraborti S, Dimitrov DS, Anderson WF. Rozenberg-Adler Y, et al. Exp Mol Pathol. 2008 Feb;84(1):18-30. doi: 10.1016/j.yexmp.2007.11.001. Epub 2007 Dec 3. Exp Mol Pathol. 2008. PMID: 18222422 - Mutational analysis of the putative receptor-binding domain of Moloney murine leukemia virus glycoprotein gp70.
Panda BR, Kingsman SM, Kingsman AJ. Panda BR, et al. Virology. 2000 Jul 20;273(1):90-100. doi: 10.1006/viro.2000.0397. Virology. 2000. PMID: 10891411 - Structural Insights into Membrane Fusion Mediated by Convergent Small Fusogens.
Yang Y, Margam NN. Yang Y, et al. Cells. 2021 Jan 15;10(1):160. doi: 10.3390/cells10010160. Cells. 2021. PMID: 33467484 Free PMC article. Review. - [Mechanisms of virus-induced membrane and cell fusion reactions].
Asano A, Asano K. Asano A, et al. Uirusu. 1988 Jun;38(1):15-22. doi: 10.2222/jsv.38.15. Uirusu. 1988. PMID: 2853482 Review. Japanese. No abstract available.
Cited by
- De novo design of conformationally flexible transmembrane peptides driving membrane fusion.
Hofmann MW, Weise K, Ollesch J, Agrawal P, Stalz H, Stelzer W, Hulsbergen F, de Groot H, Gerwert K, Reed J, Langosch D. Hofmann MW, et al. Proc Natl Acad Sci U S A. 2004 Oct 12;101(41):14776-81. doi: 10.1073/pnas.0405175101. Epub 2004 Sep 29. Proc Natl Acad Sci U S A. 2004. PMID: 15456911 Free PMC article. - Role of the specific amino acid sequence of the membrane-spanning domain of human immunodeficiency virus type 1 in membrane fusion.
Miyauchi K, Komano J, Yokomaku Y, Sugiura W, Yamamoto N, Matsuda Z. Miyauchi K, et al. J Virol. 2005 Apr;79(8):4720-9. doi: 10.1128/JVI.79.8.4720-4729.2005. J Virol. 2005. PMID: 15795258 Free PMC article. - Fv-4: identification of the defect in Env and the mechanism of resistance to ecotropic murine leukemia virus.
Taylor GM, Gao Y, Sanders DA. Taylor GM, et al. J Virol. 2001 Nov;75(22):11244-8. doi: 10.1128/JVI.75.22.11244-11248.2001. J Virol. 2001. PMID: 11602766 Free PMC article. - The transmembrane domain and cytoplasmic tail of herpes simplex virus type 1 glycoprotein H play a role in membrane fusion.
Harman A, Browne H, Minson T. Harman A, et al. J Virol. 2002 Nov;76(21):10708-16. doi: 10.1128/jvi.76.21.10708-10716.2002. J Virol. 2002. PMID: 12368313 Free PMC article. - Ross River virus glycoprotein-pseudotyped retroviruses and stable cell lines for their production.
Sharkey CM, North CL, Kuhn RJ, Sanders DA. Sharkey CM, et al. J Virol. 2001 Mar;75(6):2653-9. doi: 10.1128/JVI.75.6.2653-2659.2001. J Virol. 2001. PMID: 11222688 Free PMC article.
References
- Bedgood RM, Stallcup MR. A novel intermediate in processing of murine leukemia virus envelope glycoproteins. Proteolytic cleavage in the late Golgi region. J Biol Chem. 1992;267:7060–7065. - PubMed
- Bullough PA, Hughson FM, Skehel JJ, Wiley DC. Structure of influenza hemagglutinin at the pH of membrane fusion. Nature. 1994;371:37–43. - PubMed
- Carr CM, Kim PS. A spring-loaded mechanism for the conformational change of influenza hemagglutinin. Cell. 1993;73:823–832. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources