Neuronal expression of neural nitric oxide synthase (nNOS) protein is suppressed by an antisense RNA transcribed from an NOS pseudogene - PubMed (original) (raw)
Neuronal expression of neural nitric oxide synthase (nNOS) protein is suppressed by an antisense RNA transcribed from an NOS pseudogene
S A Korneev et al. J Neurosci. 1999.
Abstract
Here, we show that a nitric oxide synthase (NOS) pseudogene is expressed in the CNS of the snail Lymnaea stagnalis. The pseudo-NOS transcript includes a region of significant antisense homology to a previously reported neuronal NOS (nNOS)-encoding mRNA. This suggested that the pseudo-NOS transcript acts as a natural antisense regulator of nNOS protein synthesis. In support of this, we show that both the nNOS-encoding and the pseudo-NOS transcripts are coexpressed in giant identified neurons (the cerebral giant cells) in the cerebral ganglion. Moreover, reverse transcription-PCR experiments on RNA isolated from the CNS establish that stable RNA-RNA duplex molecules do form between the two transcripts in vivo. Using an in vitro translation assay, we further demonstrate that the antisense region of the pseudogene transcript prevents the translation of nNOS protein from the nNOS-encoding mRNA. By analyzing NOS RNA and nNOS protein expression in two different identified neurons, we find that when both the nNOS-encoding and the pseudo-NOS transcripts are present in the same neuron, nNOS enzyme activity is substantially suppressed. Importantly, these results show that a natural antisense mechanism can mediate the translational control of nNOS expression in the Lymnaea CNS. Our findings also suggest that transcribed pseudogenes are not entirely without purpose and are a potential source of a new class of regulatory gene in the nervous system.
Figures
Fig. 1.
Molecular cloning of the pseudo-NOS transcript.A, Sequence of a full-length cDNA clone isolated from a_Lymnaea_ CNS cDNA library. The antisense region from 93 to 238 bp is indicated in bold type. The core region of high homology (>80%) to the nNOS-encoding transcript is_shaded_. A polyadenylation signal is_underlined_. Stop codons within the core region are marked by circles: white, frame 1;shaded, frame 2; and black, frame 3.B, Schematic representation of the pseudo-NOS and nNOS-encoding transcripts. The antisense region in the pseudo-NOS transcript and its complementary counterpart are shown by_black_ and hatched boxes, respectively. Regions of high homology are shaded, and the unfilled areas have no significant homology to one another. The positions of the numbered primers used in RT-PCR experiments on isolated identified neurons are shown by arrows.
Fig. 2.
Alignment of the antisense region (93–238 nt) of the pseudo-NOS transcript with its complementary counterpart in the nNOS mRNA. In this alignment, there is ∼80% complementarity. The non-Watson–Crick G–U base pairs that are common in RNA secondary structure are shown by dots. The positions of three primers used in the identification of RNA–RNA duplexes are_underlined_ and named. Primers in positions expected to be protected from ribonuclease A (within the proposed duplex) are P2 and P3. The primer located outside of the protected area is called P1. Further explanation of the ribonuclease A protection experiment is provided in Figure 5. Details of procedures are in Materials and Methods.
Fig. 3.
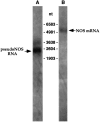
The pseudo-NOS and nNOS-encoding transcripts are expressed in the Lymnaea CNS. Northern blot analysis of_Lymnaea_ CNS poly(A+) RNA using a probe specifically recognizing the antisense region of the pseudo-NOS transcript identifies a prominent band of the expected size (∼2500 nt) in lane A (arrow). A less prominent transcript of ∼3200 nt is also revealed in the experiment. This suggests that there are other RNA molecules in the CNS that are antisense to the nNOS-encoding transcript. In lane B, the result of the hybridization with a probe recognizing the 3′ untranslated end of nNOS mRNA is shown. As expected, a single transcript of ∼5000 nt is revealed (arrow).
Fig. 4.
A uniquely identified neuron (the CGC) coexpresses functional NOS mRNA and pseudo-NOS RNA. In_A_, the results of RT-PCR experiments on RNA purified from isolated identified CGCs are illustrated. Lane 4_shows that a PCR product of the expected size (598 bp) is generated by primers specific to nNOS mRNA. Similarly, in lane 2, a PCR product of the expected size (431 bp) generated from the same RNA sample by primers specific to the pseudo-NOS RNA is detected.Lanes 1 and 3 show the results of PCR experiments designed to control for possible DNA contamination of the samples analyzed in lanes 2 and 4, respectively. In these experiments, reverse transcriptase was omitted, and as a consequence, no products were generated. In_B, the results from isolated B2 motoneurons are presented. Lane 4 shows a PCR product of the expected size (598 bp) generated by the same nNOS-specific primers as used in_A_. Note that there is no PCR product in lane 2 in which the pseudo-NOS RNA-specific primers were used.Lanes 1 and 3 represent control experiments in which reverse transcriptase was omitted. The absence of any PCR products in these lanes proves that the RNA sample used in the RT-PCR experiments was free from DNA contamination. All RT-PCR products shown have been cloned and sequenced to confirm their identity.
Fig. 5.
A schematic diagram showing the major steps of the ribonuclease protection procedure used to detect RNA–RNA duplexes_in vivo_. To preserve possible RNA–RNA hybrids, cytoplasmic RNA was purified from the CNS under nondenaturing conditions. To identify our hypothesized RNA–RNA duplex, the RNA has to be treated with RNase A, an enzyme that cleaves single-stranded but not double-stranded RNA molecules. After RNase A treatment, reverse transcription reactions in the presence of either P2 primer (located within the protected area) or P1 primer (located outside the protected area) are performed. After adding the P3 primer, a cDNA generated in the first reaction could be amplified using PCR and then will be revealed as a single band of the expected size by electrophoresis. In contrast, no cDNA could be produced in the second reverse transcription reaction, and subsequently, no PCR product is expected. In the_left column_, the predicted results of the control experiments (no RNase A treatment) are summarized. Two RT-PCR products should be detected in the reverse transcription reaction: one generated by P2 and P3 and the other by P1 and P3.
Fig. 6.
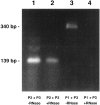
The predicted RNA–RNA duplex exists in the CNS. The experiment was performed according to the procedure described in Figure 5. Lanes 1 and 3 show the products of RT-PCR generated by RNA treated with DNase only. Lanes 2 and 4 show the products of RT-PCR generated on RNA treated with both DNase and RNase A. In lanes 1 and_2_, RNA was reverse transcribed with the P2 primer located within the predicted duplex and then amplified in the presence of the P2 and P3 primers (see Figs. 3 and 5 for location of the primers). Lanes 3 and 4 show the results generated when RNA was reverse transcribed with the P1 primer located outside the predicted duplex and then amplified in the presence of P1 and P3 primers. A product of the same predicted size (139 bp) is generated by the RNA sample that was treated with DNase only and with DNase plus RNase A (lanes 1 and 2). The band shown in lane 3 is the size predicted (340 bp) of a product generated by the P1 and P3 primers. Note that there is no product in lane 4. Importantly, these results correspond precisely to those predicted in Figure 5. All of the RT-PCR products shown have been cloned and sequenced to confirm their identity.
Fig. 7.
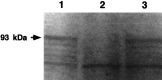
Synthesis of nNOS protein _in vitro_is suppressed by the antisense pseudo-NOS RNA. Lane 1_represents the result of the translation of NOS cRNA and shows the main labeled product is a protein of the expected (93 kDa) size (arrow). Lane 2 illustrates the effect of incubating the 0.2 μg of NOS cRNA with 2 μg of antisense pseudo-NOS cRNA before translation. Note strong suppression of translation of the nNOS protein. As a control for any effects on translation that are not related to the formation of a duplex, we also preincubated the 0.2 μg of NOS cRNA with a 2 μg of a sense version of the pseudo-NOS cRNA (lane 3). No inhibition of NOS protein synthesis is observed. Similar results, although weaker, have been obtained, even when the ratio between nNOS cRNA and pseudo-NOS transcripts was 2.5:1. A second major protein of ∼50 kDa can be seen in each_lane. This protein is present in the cell-free wheat germ system and performs a useful function as an internal control. Note that it is not diminished in intensity in lane 2.
Fig. 8.
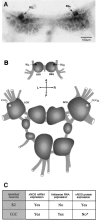
Antisense RNA-mediated regulation of nNOS protein expression in vivo. A, NADPH-diaphorase staining of the buccal ganglia. A pair of symmetrical NOS-containing diaphorase-positive B2 motoneurons is indicated by_arrows_. B, A diagram of the_Lymnaea_ CNS showing the positions of identified neurons referred to in this investigation. Dark cell bodies of B2 motoneurons reflect the fact that the neurons are strongly and consistently NADPH-diaphorase-positive, i.e., they always contain an active nNOS protein (A, C). In contrast, <10% of all CGCs examined showed nNOS enzyme activity. To emphasize this result, the cell bodies of CGCs are shown in light color. C, A summary of our experiments on the expression of nNOS mRNA, antisense RNA, and nNOS protein in the identified neurons B2 and CGC. The asterisk indicates that nNOS protein in the CGC is not entirely absent but is detected only in ∼10% of the cells observed (n = 60).
Fig. 9.
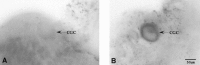
NADPH-diaphorase staining of the cerebral ganglia.A, The majority of CGCs shows no NADPH-diaphorase activity. B, One of the very few clearly NADPH-diaphorase-positive CGCs. Cell bodies of the CGCs are indicated by arrows.
Fig. 10.
A proposed model for the evolution of NOS pseudogene function.
Similar articles
- Timed and targeted differential regulation of nitric oxide synthase (NOS) and anti-NOS genes by reward conditioning leading to long-term memory formation.
Korneev SA, Straub V, Kemenes I, Korneeva EI, Ott SR, Benjamin PR, O'Shea M. Korneev SA, et al. J Neurosci. 2005 Feb 2;25(5):1188-92. doi: 10.1523/JNEUROSCI.4671-04.2005. J Neurosci. 2005. PMID: 15689555 Free PMC article. - Molecular characterization of NOS in a mollusc: expression in a giant modulatory neuron.
Korneev SA, Piper MR, Picot J, Phillips R, Korneeva EI, O'Shea M. Korneev SA, et al. J Neurobiol. 1998 Apr;35(1):65-76. doi: 10.1002/(sici)1097-4695(199804)35:1<65::aid-neu6>3.0.co;2-9. J Neurobiol. 1998. PMID: 9552167 - Partial cloning of constitutive and inducible nitric oxide synthases and detailed neuronal expression of NOS mRNA in the cerebellum and optic tectum of adult Atlantic salmon (Salmo salar).
Oyan AM, Nilsen F, Goksøyr A, Holmqvist B. Oyan AM, et al. Brain Res Mol Brain Res. 2000 May 31;78(1-2):38-49. doi: 10.1016/s0169-328x(00)00066-8. Brain Res Mol Brain Res. 2000. PMID: 10891583 - Neuronal NOS: gene structure, mRNA diversity, and functional relevance.
Wang Y, Newton DC, Marsden PA. Wang Y, et al. Crit Rev Neurobiol. 1999;13(1):21-43. doi: 10.1615/critrevneurobiol.v13.i1.20. Crit Rev Neurobiol. 1999. PMID: 10223522 Review. - Regulation of neuronal nitric oxide synthase through alternative transcripts.
Brenman JE, Xia H, Chao DS, Black SM, Bredt DS. Brenman JE, et al. Dev Neurosci. 1997;19(3):224-31. doi: 10.1159/000111211. Dev Neurosci. 1997. PMID: 9208206 Review.
Cited by
- pseudoMap: an innovative and comprehensive resource for identification of siRNA-mediated mechanisms in human transcribed pseudogenes.
Chan WL, Yang WK, Huang HD, Chang JG. Chan WL, et al. Database (Oxford). 2013 Feb 8;2013:bat001. doi: 10.1093/database/bat001. Print 2013. Database (Oxford). 2013. PMID: 23396300 Free PMC article. - Frame disruptions in human mRNA transcripts, and their relationship with splicing and protein structures.
Harrison P, Yu Z. Harrison P, et al. BMC Genomics. 2007 Oct 15;8:371. doi: 10.1186/1471-2164-8-371. BMC Genomics. 2007. PMID: 17937804 Free PMC article. - Systematic identification of pseudogenes through whole genome expression evidence profiling.
Yao A, Charlab R, Li P. Yao A, et al. Nucleic Acids Res. 2006;34(16):4477-85. doi: 10.1093/nar/gkl591. Epub 2006 Aug 31. Nucleic Acids Res. 2006. PMID: 16945953 Free PMC article. - Pseudogene: lessons from PCR bias, identification and resurrection.
Chen SM, Ma KY, Zeng J. Chen SM, et al. Mol Biol Rep. 2011 Aug;38(6):3709-15. doi: 10.1007/s11033-010-0485-4. Epub 2010 Nov 30. Mol Biol Rep. 2011. PMID: 21116863
References
- Bredt DS, Snyder SH. Nitric oxide, a novel neuronal messenger. Neuron. 1992;8:3–11. - PubMed
- Bredt DS, Hwang PM, Snyder SH. Localization of nitric oxide synthase indicating a neural role for nitric oxide. Nature. 1990;347:768–770. - PubMed
- Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials