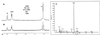Salmonella typhimurium LT2 catabolizes propionate via the 2-methylcitric acid cycle - PubMed (original) (raw)
Salmonella typhimurium LT2 catabolizes propionate via the 2-methylcitric acid cycle
A R Horswill et al. J Bacteriol. 1999 Sep.
Abstract
We previously identified the prpBCDE operon, which encodes catabolic functions required for propionate catabolism in Salmonella typhimurium. Results from (13)C-labeling experiments have identified the route of propionate breakdown and determined the biochemical role of each Prp enzyme in this pathway. The identification of catabolites accumulating in wild-type and mutant strains was consistent with propionate breakdown through the 2-methylcitric acid cycle. Our experiments demonstrate that the alpha-carbon of propionate is oxidized to yield pyruvate. The reactions are catalyzed by propionyl coenzyme A (propionyl-CoA) synthetase (PrpE), 2-methylcitrate synthase (PrpC), 2-methylcitrate dehydratase (probably PrpD), 2-methylisocitrate hydratase (probably PrpD), and 2-methylisocitrate lyase (PrpB). In support of this conclusion, the PrpC enzyme was purified to homogeneity and shown to have 2-methylcitrate synthase activity in vitro. (1)H nuclear magnetic resonance spectroscopy and negative-ion electrospray ionization mass spectrometry identified 2-methylcitrate as the product of the PrpC reaction. Although PrpC could use acetyl-CoA as a substrate to synthesize citrate, kinetic analysis demonstrated that propionyl-CoA is the preferred substrate.
Figures
FIG. 1
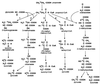
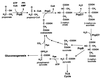
Proposed propionate breakdown pathways in E. coli and S. typhimurium. Pathways: 1, α-hydroxyglutarate; 2, citramalate; 3, methylmalonyl-CoA; 4, acryloyl-CoA; 5, 2-methylcitric acid cycle.
FIG. 2
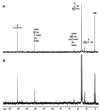
Propionate catabolites visualized by 13C-NMR spectroscopy. Strain TR6583 (prpBCDE+) was grown in minimal medium supplemented with [2-13C]propionate to accumulate 13C-labeled propionate breakdown intermediates. (A) Proton-decoupled 13C-NMR spectrum of an extract from this culture. (B) Proton-coupled 13C-NMR spectrum of this extract. The boxed carbon atoms in the chemical structures above the peaks indicate the source of the carbon signals.
FIG. 3
In vitro conversion of [2-13C]propionate to [2-13C]propionyl-CoA by PrpE. Dialyzed cell extracts of strain JE4184 were used in these experiments. (A) Proton-decoupled 13C-NMR spectrum of the reaction products. (B) Proton-coupled 13C-NMR spectrum of the reaction products.
FIG. 4
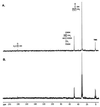
Propionate catabolites that accumulate in a prpD mutant. Strain JE3914 (prpD174) was grown in minimal medium supplemented with [2-13C]propionate to accumulate 13C-labeled propionate breakdown intermediates. (A) Proton-decoupled spectrum of an extract from this culture. (B) Proton-coupled spectrum of this extract. The boxed carbon atoms in the chemical structures above the peaks indicate the source of the carbon signals.
FIG. 5
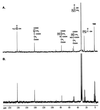
Propionate catabolites that accumulate in a prpB mutant. Strain JE3946 (prpB195) was grown in minimal medium supplemented with [2-13C]propionate to accumulate 13C-labeled propionate breakdown intermediates. (A) Proton-decoupled spectrum of an extract from this culture. (B) Proton-coupled spectrum of this extract. The boxed carbon atoms in the chemical structures above the peaks indicate the source of the carbon signals.
FIG. 6
Spectroscopic analysis of the PrpC reaction product. (A) 1H-NMR spectrum; (B) homodecoupling 1H-NMR spectrum with irradiation at the methyl doublet; (C) ESI mass spectrum. The structure shown is 2-methylcitrate.
FIG. 7
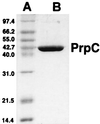
SDS-PAGE analysis of homogeneous PrpC. Lanes: A, molecular mass standards (in decreasing mass order): phosphorylase b, bovine serum albumin, glutamate dehydrogenase, ovalbumin, aldolase, carbonic anhydrase, lysozyme; B, His-tagged PrpC (3 μg) (approximately 45-kDa monomer).
FIG. 8
Pathway for propionate catabolism in S. typhimurium. The results presented in this paper support the breakdown of propionate via 2-methylcitrate. The reactions catalyzed by enzymes encoded in the prpBCDE operon are indicated. A possible route for oxaloacetate regeneration and gluconeogenesis is shown (pps, PEP synthetase; Ppc, PEP carboxylase).
FIG. 9
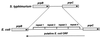
Comparison of the prpBC intergenic region between S. typhimurium and E. coli. The 91-bp repeats and proposed ORF in E. coli are indicated.
Similar articles
- Oxidation of propionate to pyruvate in Escherichia coli. Involvement of methylcitrate dehydratase and aconitase.
Brock M, Maerker C, Schütz A, Völker U, Buckel W. Brock M, et al. Eur J Biochem. 2002 Dec;269(24):6184-94. doi: 10.1046/j.1432-1033.2002.03336.x. Eur J Biochem. 2002. PMID: 12473114 - The Methylcitrate Cycle and Its Crosstalk with the Glyoxylate Cycle and Tricarboxylic Acid Cycle in Pathogenic Fungi.
Huang Z, Wang Q, Khan IA, Li Y, Wang J, Wang J, Liu X, Lin F, Lu J. Huang Z, et al. Molecules. 2023 Sep 17;28(18):6667. doi: 10.3390/molecules28186667. Molecules. 2023. PMID: 37764443 Free PMC article. Review. - Loving the poison: the methylcitrate cycle and bacterial pathogenesis.
Dolan SK, Wijaya A, Geddis SM, Spring DR, Silva-Rocha R, Welch M. Dolan SK, et al. Microbiology (Reading). 2018 Mar;164(3):251-259. doi: 10.1099/mic.0.000604. Epub 2018 Jan 22. Microbiology (Reading). 2018. PMID: 29458664 Review.
Cited by
- The intestinal fatty acid propionate inhibits Salmonella invasion through the post-translational control of HilD.
Hung CC, Garner CD, Slauch JM, Dwyer ZW, Lawhon SD, Frye JG, McClelland M, Ahmer BM, Altier C. Hung CC, et al. Mol Microbiol. 2013 Mar;87(5):1045-60. doi: 10.1111/mmi.12149. Epub 2013 Jan 28. Mol Microbiol. 2013. PMID: 23289537 Free PMC article. - Short-chain fatty acid activation by acyl-coenzyme A synthetases requires SIR2 protein function in Salmonella enterica and Saccharomyces cerevisiae.
Starai VJ, Takahashi H, Boeke JD, Escalante-Semerena JC. Starai VJ, et al. Genetics. 2003 Feb;163(2):545-55. doi: 10.1093/genetics/163.2.545. Genetics. 2003. PMID: 12618394 Free PMC article. - prpR, ntrA, and ihf functions are required for expression of the prpBCDE operon, encoding enzymes that catabolize propionate in Salmonella enterica serovar typhimurium LT2.
Palacios S, Escalante-Semerena JC. Palacios S, et al. J Bacteriol. 2000 Feb;182(4):905-10. doi: 10.1128/JB.182.4.905-910.2000. J Bacteriol. 2000. PMID: 10648513 Free PMC article. - Characterization of a Glycyl Radical Enzyme Bacterial Microcompartment Pathway in Rhodobacter capsulatus.
Schindel HS, Karty JA, McKinlay JB, Bauer CE. Schindel HS, et al. J Bacteriol. 2019 Feb 11;201(5):e00343-18. doi: 10.1128/JB.00343-18. Print 2019 Mar 1. J Bacteriol. 2019. PMID: 30510145 Free PMC article. - Biosynthesis of polyketide synthase extender units.
Chan YA, Podevels AM, Kevany BM, Thomas MG. Chan YA, et al. Nat Prod Rep. 2009 Jan;26(1):90-114. doi: 10.1039/b801658p. Nat Prod Rep. 2009. PMID: 19374124 Free PMC article. Review.
References
- Aoki H, Uchiyama H, Umetsu H, Tabuchi T. Isolation of 2-methylcitrate dehydratase, a new enzyme serving in the methylcitric acid cycle for propionate metabolism, from Yallowia lipolytica. Biosci Biotechnol Biochem. 1995;59:1825–1828.
- Ausubel F A, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: Greene Publishing Associates & Wiley Interscience; 1989.
- Blair J M. Magnesium and aconitase equilibrium: determination of apparent stability constants of magnesium substrate complexes from equilibrium data. Eur J Biochem. 1969;8:287–291. - PubMed
- Blattner F R, Plunkett III G, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, Gregor J, Davis N W, Kirkpatrick H A, Goeden M A, Rose D J, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. - PubMed
- Chan R K, Botstein D, Watanabe T, Ogata Y. Specialized transduction of tetracycline resistance by phage P22 in Salmonella typhimurium. II. Properties of a high transducing lysate. Virology. 1972;50:883–898. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P41 RR002301/RR/NCRR NIH HHS/United States
- R01 GM062203/GM/NIGMS NIH HHS/United States
- GM08349/GM/NIGMS NIH HHS/United States
- T32 GM008349/GM/NIGMS NIH HHS/United States
- RR02301/RR/NCRR NIH HHS/United States
- S10 RR002781/RR/NCRR NIH HHS/United States
- S10 RR008438/RR/NCRR NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials