Search for the mechanism of genetic variation in the pro gene of human immunodeficiency virus - PubMed (original) (raw)
Search for the mechanism of genetic variation in the pro gene of human immunodeficiency virus
I M Rouzine et al. J Virol. 1999 Oct.
Abstract
To study the mechanism of evolution of the human immunodeficiency virus (HIV) protease gene (pro), we analyzed a database of 213 pro sequences isolated from 11 HIV type 1-infected patients who had not been treated with protease inhibitors. Variation in pro is restricted to rare variable bases which are highly diverse and differ in location among individuals; an average variable base appears in about 16% of individuals. The average intrapatient distance per individual variable site, 27%, is similar for synonymous and nonsynonymous sites, although synonymous sites are twice as abundant. The latter observation excludes selection for diversity as an important, permanently acting factor in the evolution of pro and leaves purifying selection as the only kind of selection. Based on this, we developed a model of evolution, both within individuals and along the transmission chain, which explains variable sites as slightly deleterious mutants slowly reverting to the better-fit variant during individual infection. In the case of a single-source transmission, genetic bottlenecks at the moment of transmission effectively suppress selection, allowing mutants to accumulate along the transmission chain to high levels. However, even very rare coinfections from independent sources are, as we show, able to counteract the bottleneck effect. Therefore, there are two possible explanations for the high mutant frequency. First, the frequency of coinfection in the natural host population may be quite low. Alternatively, a strong variation of the best-adapted sequence between individuals could be caused by a combination of an immune response present in early infection and coselection.
Figures
FIG. 1
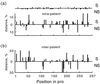
Intrapatient (a) and interpatient (b) genetic distances, averaged over patients, at different positions in pro. The upper and lower histograms in each figure correspond to synonymous and nonsynonymous sites, respectively. Dots on the upper horizontal line in panel a show the positions of sporadic mutations (seen only once in the data set).
FIG. 2
Gray-scale diagram of intrapatient genetic distance, Tki, in different patients at different variable sites in pro. The intrapatient genetic distance is indicated by the degree of shading, as shown on the scale on the right. Letters and numbers under the diagram show consensus nucleotides and positions in pro, respectively.
FIG. 3
Model of evolution of a nucleotide along the chain of infected individuals under purifying selection. The numbers at the top denote cycles of transmission. X signs denote mutants. A mutant base appears spontaneously in person n, who infects person A with the mutant and person B with the wild-type variant. Person A passes the mutant down the chain to person C, after which his/her own virus population slowly reverts to the wild type. In person D, who is stably coinfected with a pair of sequences polymorphous at that base, selection clears the mutant virus rapidly.
FIG. 4
Time dependence of the mutant frequency at different initial populations: purely mutant (reversion) (thicker line in upper panel), purely wild type (accumulation) (lower panel), or 50% mutant (growth competition) (dashed line). The thinner curve shows the intrapatient genetic distance T(t) during reversion. The horizontal bar is the time interval during which the base would be classified as variable (5 to 95%). Values shown in the upper left corner correspond to parameters as follows: the forward (wild type → mutant) and reverse mutation rates, μf and μr, corresponding to either A or C wild type; the replication cycle time, _t_rep; and selection coefficient, s. The reversion half-time, _t_50, is given by the equation _t_50 = (_t_rep/s)ln(s/μr).
FIG. 5
(a) Probability of a mutant base being in the inoculum at the chain steady state, f*∞, as a function of the transmission time, _t_∗. Solid line, strictly single-clone infection; dashed line, coinfection from two sources with probability q = 0.05. (b) Number of cycles required to reach the chain steady state, _n_eq, as a function of transmission time, _t_∗. (c) Probability of a mutant base for a very short transmission time, _t_∗ → 0, versus the probability of dual infection, q. Upper and lower curves correspond to wild-type A/C and G/T, respectively. (d) The same probability versus s, for a fixed value q = 0.01. _t_rep = 2 days; μf = 4 · 10−5, μr = 4 · 10−6 for wild type A/C and vice versa for G/T.
FIG. 6
A working model of evolution in pro. A quick initial switch of dominant epitopes redefines wild type for a number of sites, which then revert to this new wild type at speeds inversely proportional to their selection coefficients. (a) Time dependence of mutant frequency at three sites linked to an epitope. The three selection coefficients after the epitope switch are shown near corresponding curves. (b) Evolution of an individual consensus sequence at the three sites. pt, patient.
Similar articles
- Diversity of human immunodeficiency virus type 1 subtype A and CRF03_AB protease in Eastern Europe: selection of the V77I variant and its rapid spread in injecting drug user populations.
Roudinskii NI, Sukhanova AL, Kazennova EV, Weber JN, Pokrovsky VV, Mikhailovich VM, Bobkov AF. Roudinskii NI, et al. J Virol. 2004 Oct;78(20):11276-87. doi: 10.1128/JVI.78.20.11276-11287.2004. J Virol. 2004. PMID: 15452247 Free PMC article. - Evolution of pathogenic viruses with special reference to the rates of synonymous and nonsynonymous substitutions.
Gojobori T, Yamaguchi Y, Ikeo K, Mizokami M. Gojobori T, et al. Jpn J Genet. 1994 Oct;69(5):481-8. doi: 10.1266/jjg.69.481. Jpn J Genet. 1994. PMID: 7999369 Review. - Constraints on the sequence diversity of the protease of human immunodeficiency virus type 1: a guide for drug design.
Kaplan AH. Kaplan AH. AIDS Res Hum Retroviruses. 1996 Jul 1;12(10):849-53. doi: 10.1089/aid.1996.12.849. AIDS Res Hum Retroviruses. 1996. PMID: 8798968 Review. No abstract available.
Cited by
- The M184V mutation in reverse transcriptase can delay reversion of attenuated variants of simian immunodeficiency virus.
Whitney JB, Oliveira M, Detorio M, Guan Y, Wainberg MA. Whitney JB, et al. J Virol. 2002 Sep;76(17):8958-62. doi: 10.1128/jvi.76.17.8958-8962.2002. J Virol. 2002. PMID: 12163615 Free PMC article. - High-resolution molecular epidemiology and evolutionary history of HIV-1 subtypes in Albania.
Salemi M, de Oliveira T, Ciccozzi M, Rezza G, Goodenow MM. Salemi M, et al. PLoS One. 2008 Jan 2;3(1):e1390. doi: 10.1371/journal.pone.0001390. PLoS One. 2008. PMID: 18167549 Free PMC article. - Estimate of effective recombination rate and average selection coefficient for HIV in chronic infection.
Batorsky R, Kearney MF, Palmer SE, Maldarelli F, Rouzine IM, Coffin JM. Batorsky R, et al. Proc Natl Acad Sci U S A. 2011 Apr 5;108(14):5661-6. doi: 10.1073/pnas.1102036108. Epub 2011 Mar 21. Proc Natl Acad Sci U S A. 2011. PMID: 21436045 Free PMC article. - An Evolutionary Model of Progression to AIDS.
Rouzine IM. Rouzine IM. Microorganisms. 2020 Oct 31;8(11):1714. doi: 10.3390/microorganisms8111714. Microorganisms. 2020. PMID: 33142907 Free PMC article. - The evolutionary origin of the universal distribution of mutation fitness effect.
Barlukova A, Rouzine IM. Barlukova A, et al. PLoS Comput Biol. 2021 Mar 8;17(3):e1008822. doi: 10.1371/journal.pcbi.1008822. eCollection 2021 Mar. PLoS Comput Biol. 2021. PMID: 33684109 Free PMC article.
References
- Barnes W M. The fidelity of Taq polymerase catalyzing PCR is improved by an N-terminal deletion. Gene. 1992;112:29–35. - PubMed
- Burns D P, Desrosiers R C. Envelope sequence variation, neutralizing antibodies, and primate lentivirus persistence. Curr Top Microbiol Immunol. 1994;188:185–219. . (Review.) - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources